emzed is an open source toolbox for rapid and interactive development of LCMS data analysis workflows in Python and makes experimenting with new analysis strategies for LCMS data as easy as possible.
Click on images to enlarge.
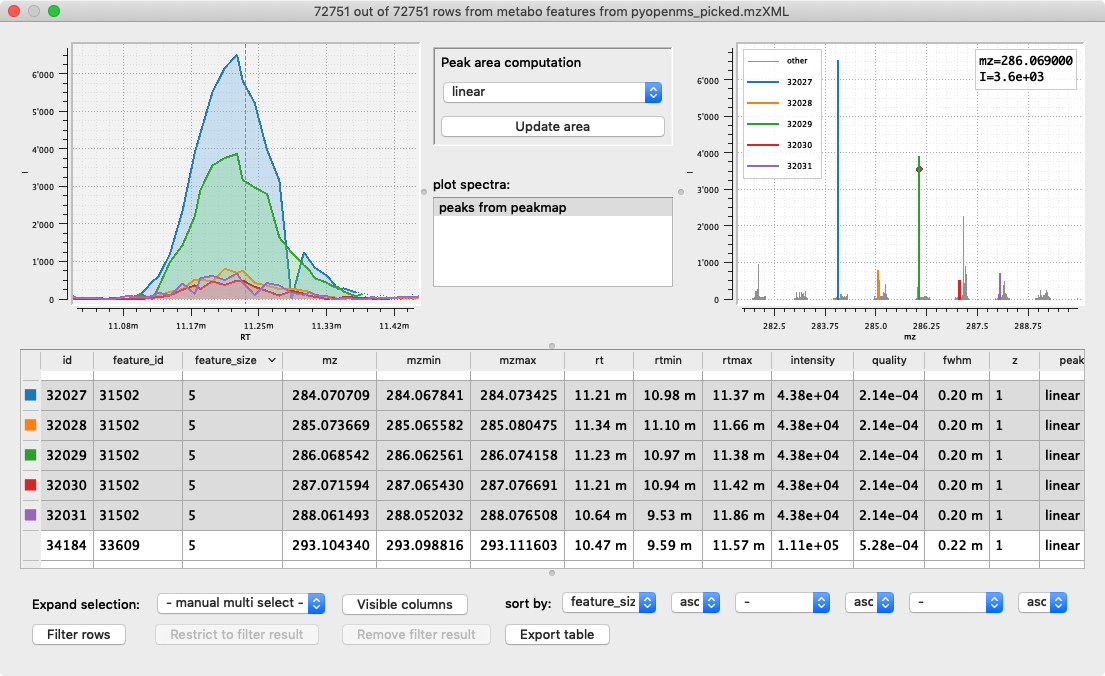
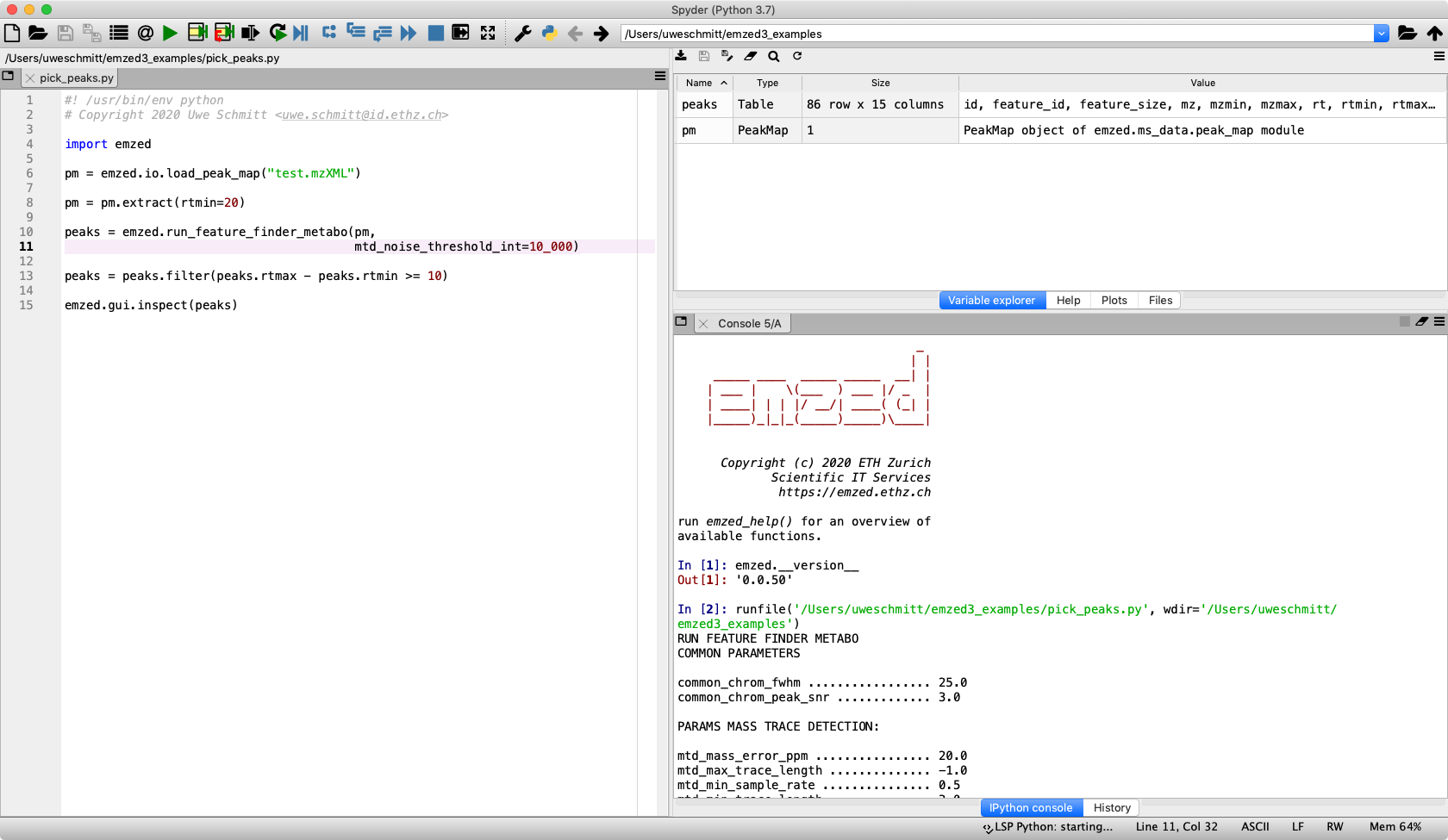
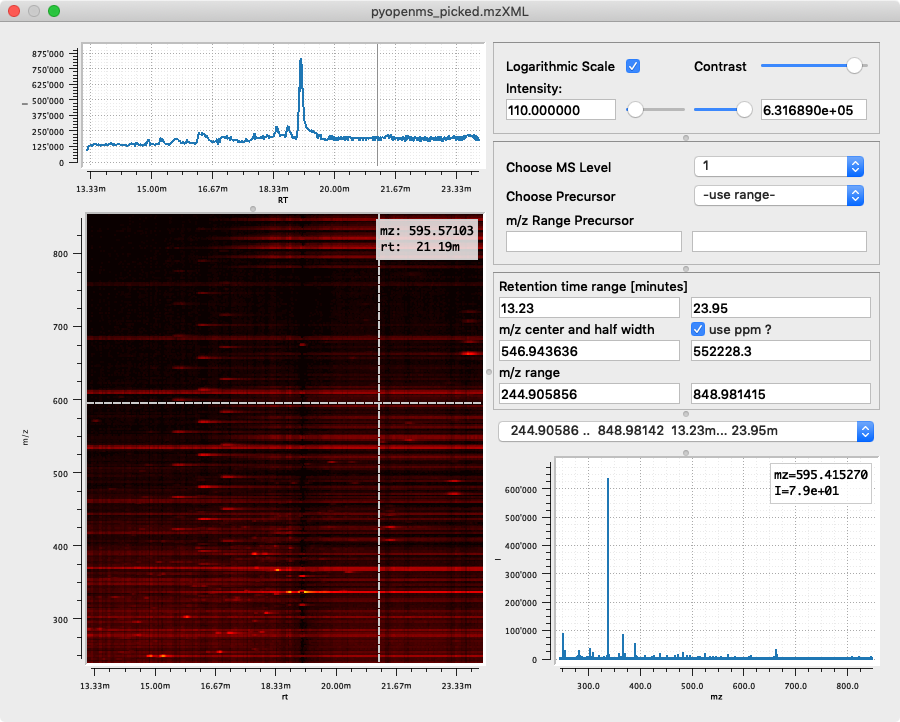
We choose Python which makes programming with emzed as simple as possible and approachable by LCMS experts without professional programming experience.
Analysis workflows consist of Python scripts composing function calls from the emzed library.
Thus processing steps are explicitly written down as Python code which is crucial for reproducible research.
In order to strengthen the trust in analysis results, emzed provides interactive
visualization tools for intermediate and final results. An emzed workflow can deliver
more insight into the achieved results than a bunch of numbers or static plots.
emzed.spyder is an integrated development environment (IDE)
based on the spyder IDE tailored to
support the overall development process.
When we started developing LCSM analysis workflows in 2012, we realized that the existing software landscape can be divided into two groups. On the one hand there are fast and flexible frameworks, but in languages like C++, which can only be used efficiently by experienced programmers. On the other hand, there are applications with graphical user interfaces that are easy to use and learn, but difficult to adapt for special needs.
Our goal was to develop a toolbox that combines the positive aspects of both groups.
The invention of programming environments such as Matlab and R leveraged
the productivity of mathematicians and scientists from other fields. emzed attempts
to introduce this concept for analyzing LCMS data.
Instead of reinventing the wheel we cherry pick algorithms from established
libraries and frameworks such as OpenMs. We
provide a consistent application programming interface (API) and
emzed based workflows avoid manual and error prone import and export
steps.
We split emzed into a user interface agnostic core library (also called
emzed) which can be used on display less servers such as high performance
computers, a library for the interactive inspection of LCMS data structures emzed.gui
and the spyder based development environment emzed.spyder.
emzed can be extended by community extensions, right now we offer an extension
emzed.ext.mzmine2 which wraps some algorithms from MZmine2.
Recent publications using emzed for data analysis:
Bidong D. Nguyen et al. Import of Aspartate and Malate by DcuABC Drives H2/Fumarate Respiration to Promote Initial Salmonella Gut-Lumen Colonization in Mice. Cell Host & Microbe (2020), https://doi.org/10.1016/j.chom.2020.04.013
Hartl et al. Untargeted metabolomics links glutathione to bacterial cell cycle progression. Nature Metabolism (2020), https://doi.org/10.1038/s42255-019-0166-0
Hartl et al. Untargeted metabolomics links glutathione to bacterial cell cycle progression, Nature Metabolism (2020), https://doi.org/10.1038/s42255-019-0166-0
Hemmann et al. Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites. PNAS (2019), https://doi.org/10.1073/pnas.1911595116
For the full list of publications continue to read here.
Original publication:
Kiefer Patrick, Schmitt Uwe, Vorholt Julia. eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows, Bioinformatics, Volume 29, Issue 7, 1 April 2013, Pages 963–964, https://doi.org/10.1093/bioinformatics/btt080
© Copyright 2012-2024 ETH Zurich
Last build 2024-03-25 10:41:42.995953.
Created using Sphinx 7.2.6.