[1]:
%%capture
# Setup
import emzed
import os
# path to files
data_folder = os.path.join(os.getcwd(), "tutorial_data")
path_sample_caulobacter = os.path.join(
data_folder, "AA_sample_caulobacter.mzML"
)
path_sample_arabidopsis = os.path.join(
data_folder, "AA_sample_arabidopsis.mzML"
)
path_caulo_final = os.path.join(data_folder, "t_adap_final.table")
path_caulo_annotated = os.path.join(
data_folder, "t_adap_annotated.table"
)
ref_path = os.path.join(data_folder, "mz_calibration_table.csv")
# peakmaps and tables
peakmap_arabidopsis = emzed.io.load_peak_map(path_sample_arabidopsis)
peakmap_caulobacter = emzed.io.load_peak_map(path_sample_caulobacter)
adap = emzed.io.load_table(path_caulo_final)
ta_caulo_annotated = emzed.io.load_table(path_caulo_annotated)
t_ref = emzed.io.load_csv(ref_path)
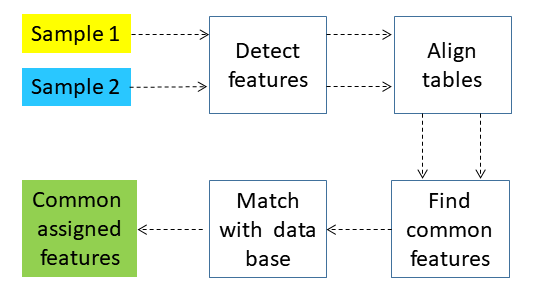
emzed3 heavily features untargeted analysis. To do so emzed3 provides tools from OpenMS and MZmine2. Whereas open ms features are integrated in the emzed3 core mzmine2 is an extension that has to be installed first. To get familiar with those tools we will exercise the basic workflow depicted above which comprises the core peak processing steps detection, alignment, and grouping. Moreover, we will perform some basic sample comparison applying data base matching with emzed integrated pubchem library.
As already mentioned above MzMine2 is an emzed extenion, and must be installed first. Open your IPython console and run the command:
emzed_install_ext('mzmine2')
After installing, close the current console and open a new one to update the installation.
Next, type the command:
emzed.ext.mzmine2.init()
Again, close the current console and start a new session. After opening the new console MZmine2 features will be available in the emzed name space
emzed.ext.mzmine2
or
mzmine2 = emzed.ext.mzmine2
A detailed user manual with all features can be find here. Note that currently, only feature detection related tools are implemented in emzed3.
Let’s start with a more general look at LC-MS data structure. LC-MS level 1 data acquisition can be regarded as a 2 dimensional separation procedure. In the first dimension, LC separates compounds by physico-chemical interactions over time (retention time). In the second dimension, compound ions are separated by their mass to charge ratios m/z whereby the continuous LC output is scanned with an MS instrument specific frequency. Hence, we can define LC-MS peaks as sequences of spectra with m/z specific intensity series of with shapes typical for applied LC separation. Accordingly, we can subdivide the task of peak detection algorithm into two principal steps:
EIC detection: Search the peak map for all series of consecutive m/z peaks with the same m/z values (EIC).
Peak detection: Find all chromatographic (LC) peaks within each EIC.
To find an EIC we have to define the the meaning of m/z peaks (signal), what means a series of consecutive m/z peaks, and what means m/z peaks with same value. To define a signal we have to distinguish between signal (S) and noise (N). Remember, it is commonn practice to the define presence of a signal by a \(S/N >=3\) and a quantifiable signal by \(S/N >=10\). The number of (consecutive) m/z peaks or the time range considered to define a m/z trace directly depends of the applied LC-method and on the scan speed of the instrument. Whereas HPLC columns produce peaks with peak widths in the range of 20-30s, UPLC peaks are up to 10x narrower. Independent on the applied column type, compound peaks of interest can strongly differ from the typical shape and hence the range must be chosen with care. Finally, the variation of measured m/z values depends on the mass resolution \(R = mz/\Delta mz\) (measured at full width half maximum, FWHM) and is not only instrument dependent but also depends on the m/z itself for most common MS instruments (TOF, Orbitrap). The width of a m/z peak can be easily seen when data were acquired in the profile mode. However, most algorithms work with centroided data, where each spectral peak is represented by its centroided value. In that case the R value of the peak or the spectrum is required. A more practical approach is to determine the observed m/z width (max(m/z) - min(m/z)) directly from the raw data:
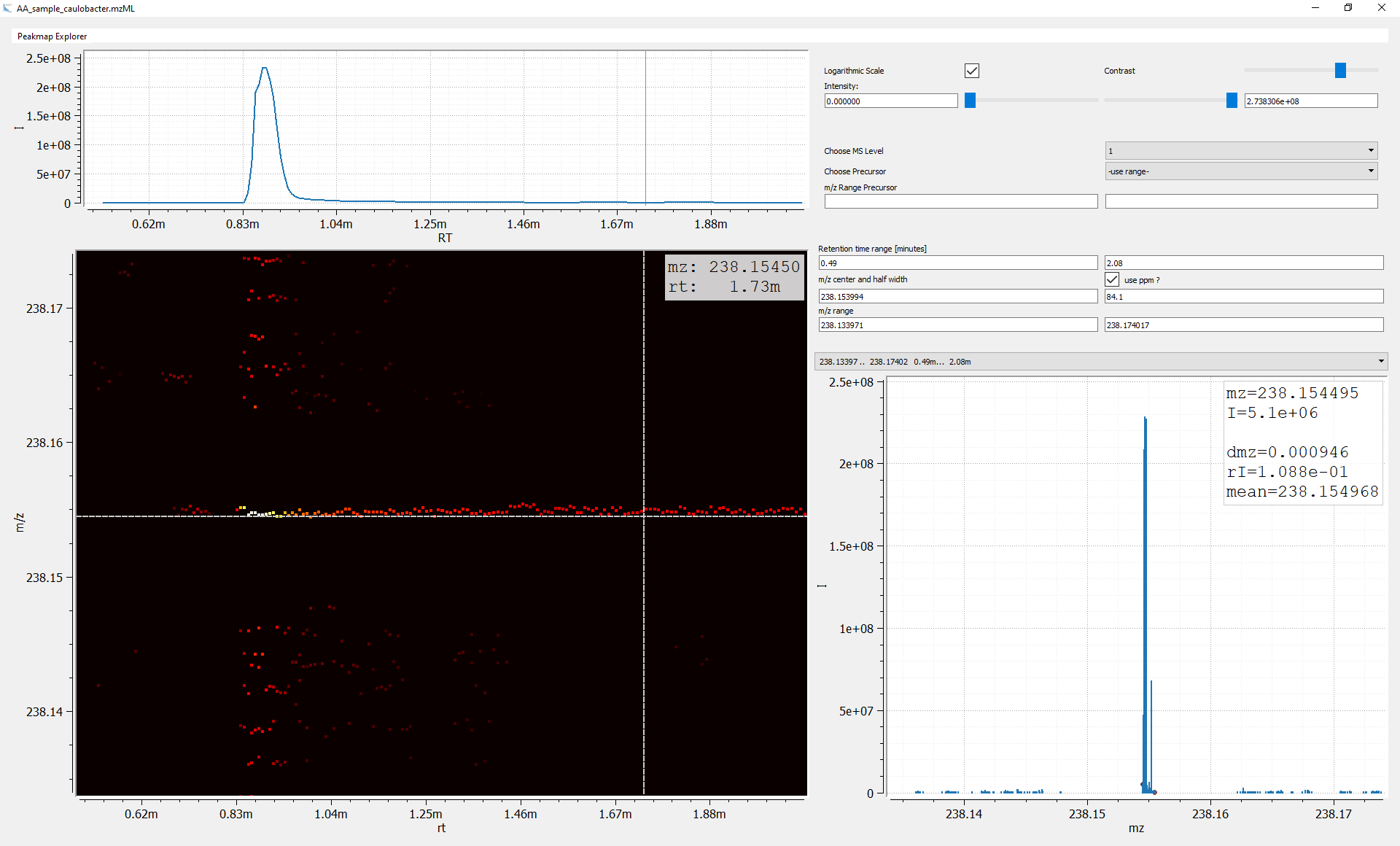
Figure 3: Measuring m/z width with PeakMap explorer using the plot of the summed spectra. For the given EIC, the m/z difference of the minimal and maximal measured m/z value is dmz = 0.000946.
Figure 3 shows a typical Orbitrap m/z trace. In this example the absolute trace width is around 1 mmU and the relative about 4 ppm (m/z = 238.154). If the algorithm requires relative values the acquisition m/z range is crucial to provide reasonable values (1 mmu at m/z 75.0 corresponds to ~13 ppm).
Once EICs are available, LC-peaks can be defined. Similar to mass trace detection, the values start, apex and end of LC peaks must be determined. Since those parameters are much more algorithm dependent, we will discuss peak detection in more details directly with provided peak detectors.
We will introduce untargeted feature detection using LC-MS data acquired with an Orbitrap MS instrument and hence, some aspects will be instrument specific.
emzed3 provides openMS run_feature_finder_metabo for feature detection. To explore the command type:
help(emzed.run_feature_finder_metabo)
run_feature_finder_metabo has a high number of function arguments and adaptation to the applied LC-MS method is not trivial. If we have a closer look at the dectection parameter names, we can assign those parameters to the different processing steps by their prefixes:
common: common parameters
mtd: mass trace detection
epdet: elution peak detection
ffm: feature finding metabo
The last process is only executed if run_feature_grouper = True and provides isotopologue peak grouping.
Note, it’s much more convenient and less error prone to use key word arguments kwargs for parameter settings. For the given data set we provide a dictionary with optimized parameters:
[2]:
kwargs = dict(
common_chrom_peak_snr=10.0,
common_chrom_fwhm=3.0,
mtd_noise_threshold_int=7000.0,
mtd_mass_error_ppm=20.0,
mtd_reestimate_mt_sd="true",
mtd_trace_termination_criterion="outlier",
mtd_trace_termination_outliers=4,
mtd_min_sample_rate=0.5,
mtd_min_trace_length=5.0,
mtd_max_trace_length=-1.0,
epdet_width_filtering="auto",
epdet_masstrace_snr_filtering="false",
ffm_local_rt_range=2.0,
ffm_local_mz_range=5.0,
ffm_charge_lower_bound=0,
ffm_charge_upper_bound=3,
ffm_report_summed_ints="false",
ffm_isotope_filtering_model="none",
ffm_use_smoothed_intensities="false",
)
We can now process both samples:
[3]:
t_ara = emzed.run_feature_finder_metabo(
peakmap_arabidopsis, verbose=False, **kwargs
)
t_caulo = emzed.run_feature_finder_metabo(
peakmap_caulobacter, verbose=False, **kwargs
)
pyopenms: pw low: 1.48233 pw high: 6.2303
pyopenms: Warning: 'report_smoothed_intensities' is set to true, but 'use_smoothed_intensities' is false. Ignoring 'report_smoothed_intensities'.
pyopenms: pw low: 1.7154 pw high: 6.51534
Note, verbose == False omits data processing output. Let’s have a look at the Caulobacter resulting table:
[4]:
t_caulo.summary()
[4]:
| id | name | type | format | nones | len | min | max | distinct values |
|---|---|---|---|---|---|---|---|---|
| int | str | str | str | int | int | float | float | int |
| 0 | id | int | %d | 0 | 1042 | 0.000000 | 1041.000000 | 1042 |
| 1 | feature_id | int | %d | 0 | 1042 | 0.000000 | 778.000000 | 779 |
| 2 | feature_size | int | %d | 0 | 1042 | 1.000000 | 4.000000 | 4 |
| 3 | mz | MzType | %11.6f | 0 | 1042 | 85.075765 | 599.389233 | 1042 |
| 4 | mzmin | MzType | %11.6f | 0 | 1042 | 85.075607 | 599.388428 | 1037 |
| 5 | mzmax | MzType | %11.6f | 0 | 1042 | 85.075829 | 599.390381 | 1039 |
| 6 | rt | RtType | rt_formatter | 0 | 1042 | 28.472400 | 401.853800 | 265 |
| 7 | rtmin | RtType | rt_formatter | 0 | 1042 | 26.869800 | 397.279000 | 290 |
| 8 | rtmax | RtType | rt_formatter | 0 | 1042 | 30.920500 | 599.716100 | 315 |
| 9 | intensity | float | %.2e | 0 | 1042 | 38101.944970 | 1174356996.343237 | 1042 |
| 10 | quality | float | %.2e | 0 | 1042 | 0.000011 | 0.295786 | 779 |
| 11 | fwhm | RtType | rt_formatter | 0 | 1042 | 1.715400 | 6.514842 | 779 |
| 12 | z | int | %d | 0 | 1042 | 0.000000 | 3.000000 | 4 |
| 13 | peakmap | PeakMap | None | 0 | 1042 | - | - | 1 |
| 14 | source | str | %s | 0 | 1042 | - | - | 1 |
With given parameters we detected 1149 peaks represented by 1149 different id values. Those peaks correspond to 863 isotopologue features grouped by feature_id. Note, id and feature_id are sample specific! Let’s print the 10 most intense peaks of the feature table:
[5]:
t_caulo.sort_by("intensity", ascending=False)[:10]
[5]:
| id | feature_id | feature_size | mz | mzmin | mzmax | rt | rtmin | rtmax | intensity | quality | fwhm | z | source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str |
| 1 | 0 | 4 | 245.149248 | 245.148941 | 245.149796 | 1.53 m | 1.41 m | 2.15 m | 1.17e+09 | 2.96e-01 | 0.09 m | 1 | AA_sample_caulobacter.mzML |
| 4 | 1 | 3 | 238.154740 | 238.154495 | 238.155441 | 0.88 m | 0.84 m | 1.51 m | 6.88e+08 | 1.74e-01 | 0.06 m | 1 | AA_sample_caulobacter.mzML |
| 2 | 0 | 4 | 246.152416 | 246.152115 | 246.153122 | 1.53 m | 1.42 m | 2.16 m | 1.43e+08 | 2.96e-01 | 0.09 m | 1 | AA_sample_caulobacter.mzML |
| 7 | 2 | 3 | 153.101914 | 153.101608 | 153.102173 | 1.00 m | 0.95 m | 1.20 m | 1.13e+08 | 2.78e-02 | 0.04 m | 1 | AA_sample_caulobacter.mzML |
| 10 | 3 | 3 | 182.164898 | 182.164536 | 182.165176 | 2.36 m | 2.26 m | 4.17 m | 1.08e+08 | 2.67e-02 | 0.10 m | 1 | AA_sample_caulobacter.mzML |
| 13 | 4 | 3 | 171.112591 | 171.112335 | 171.112778 | 1.25 m | 1.20 m | 1.42 m | 9.37e+07 | 2.30e-02 | 0.05 m | 1 | AA_sample_caulobacter.mzML |
| 5 | 1 | 3 | 239.157875 | 239.157730 | 239.158966 | 0.88 m | 0.84 m | 1.50 m | 9.03e+07 | 1.74e-01 | 0.06 m | 1 | AA_sample_caulobacter.mzML |
| 16 | 5 | 2 | 160.180614 | 160.180328 | 160.180893 | 6.23 m | 6.18 m | 10.00 m | 8.23e+07 | 2.01e-02 | 0.06 m | 1 | AA_sample_caulobacter.mzML |
| 18 | 6 | 2 | 102.127362 | 102.127266 | 102.127548 | 0.93 m | 0.90 m | 1.39 m | 6.95e+07 | 1.68e-02 | 0.04 m | 1 | AA_sample_caulobacter.mzML |
| 20 | 7 | 2 | 139.122672 | 139.122513 | 139.122894 | 0.95 m | 0.91 m | 1.44 m | 4.51e+07 | 1.10e-02 | 0.05 m | 1 | AA_sample_caulobacter.mzML |
All columns required for peak visualization and integration are provided (mzmin, mzmax, rtmin, rtmax, peakmap). The charge state of a feature is shown in column z. In case z equals 0, isotopoloque grouping is missing and no charge state could be assigned. Moreover, column intensity does not refer to the individual peak intensity but is the same for all peaks of the same feature configured by the paramter ffm_report_sum_ints ('true': sum of all peak intensity, 'false':
intensity of monoisotopopic peak).
Next, we compare the most intense feature of the Arabidopsis sample with the corresponding one in the Caulobacter sample:
[6]:
t_comp = t_ara.left_join(
t_caulo,
t_ara.mz.approx_equal(t_caulo.mz, 0.003, 0)
& t_ara.rt.approx_equal(t_caulo.rt, 20.0, 0),
)
In this example we used the left_join Table method since all rows of the reference table (the left table) are kept in the result table and missing values in the right Table are set to None. We compare peaks m/z and RT values using the Column method approx_equal(other, atol, rtol)) with
other: column of other (right) Table
atol: absolute allowed tolerance
rtol: allowed tolerance relative to value in left Table
[7]:
t_comp.summary()
[7]:
| id | name | type | format | nones | len | min | max | distinct values |
|---|---|---|---|---|---|---|---|---|
| int | str | str | str | int | int | float | float | int |
| 0 | id | int | %d | 0 | 410 | 0.000000 | 407.000000 | 408 |
| 1 | feature_id | int | %d | 0 | 410 | 0.000000 | 318.000000 | 319 |
| 2 | feature_size | int | %d | 0 | 410 | 1.000000 | 4.000000 | 4 |
| 3 | mz | MzType | %11.6f | 0 | 410 | 102.054852 | 960.576274 | 408 |
| 4 | mzmin | MzType | %11.6f | 0 | 410 | 102.054733 | 960.573914 | 405 |
| 5 | mzmax | MzType | %11.6f | 0 | 410 | 102.054993 | 960.578186 | 405 |
| 6 | rt | RtType | rt_formatter | 0 | 410 | 26.818200 | 393.331500 | 170 |
| 7 | rtmin | RtType | rt_formatter | 0 | 410 | 25.168600 | 389.327900 | 192 |
| 8 | rtmax | RtType | rt_formatter | 0 | 410 | 31.236300 | 397.905100 | 217 |
| 9 | intensity | float | %.2e | 0 | 410 | 42038.464802 | 44851637.202265 | 408 |
| 10 | quality | float | %.2e | 0 | 410 | 0.000060 | 0.067584 | 319 |
| 11 | fwhm | RtType | rt_formatter | 0 | 410 | 1.482326 | 6.230299 | 319 |
| 12 | z | int | %d | 0 | 410 | 0.000000 | 1.000000 | 2 |
| 13 | peakmap | PeakMap | None | 0 | 410 | - | - | 1 |
| 14 | source | str | %s | 0 | 410 | - | - | 1 |
| 15 | id__0 | int | %d | 384 | 410 | 65.000000 | 971.000000 | 24 |
| 16 | feature_id__0 | int | %d | 384 | 410 | 26.000000 | 708.000000 | 24 |
| 17 | feature_size__0 | int | %d | 384 | 410 | 1.000000 | 3.000000 | 3 |
| 18 | mz__0 | MzType | %11.6f | 384 | 410 | 104.070310 | 406.243648 | 24 |
| 19 | mzmin__0 | MzType | %11.6f | 384 | 410 | 104.070198 | 406.242706 | 24 |
| 20 | mzmax__0 | MzType | %11.6f | 384 | 410 | 104.070457 | 406.244476 | 24 |
| 21 | rt__0 | RtType | rt_formatter | 384 | 410 | 36.803200 | 315.126200 | 20 |
| 22 | rtmin__0 | RtType | rt_formatter | 384 | 410 | 33.335500 | 313.410600 | 22 |
| 23 | rtmax__0 | RtType | rt_formatter | 384 | 410 | 39.315900 | 342.005800 | 23 |
| 24 | intensity__0 | float | %.2e | 384 | 410 | 170555.439210 | 14589258.617420 | 24 |
| 25 | quality__0 | float | %.2e | 384 | 410 | 0.000040 | 0.003667 | 24 |
| 26 | fwhm__0 | RtType | rt_formatter | 384 | 410 | 1.923615 | 6.021642 | 24 |
| 27 | z__0 | int | %d | 384 | 410 | 0.000000 | 1.000000 | 2 |
| 28 | peakmap__0 | PeakMap | None | 384 | 410 | - | - | 1 |
| 29 | source__0 | str | %s | 384 | 410 | - | - | 1 |
The Table summary of columns id and id__0 shows that only 25 peaks of Caulobacter had a match with an Arabidopsis sample peak. Moreover, the distinct values of id is 451 whereas table length is 454 showing that 3 ambiguous matches occured. This is not very surprising since we set RT tolerance to 20 s. Finally we will check, how many of 10 most intend features have a match in Caulobacter sample:
[8]:
t_comp.sort_by("intensity")[:10]
[8]:
| id | feature_id | feature_size | mz | mzmin | mzmax | rt | rtmin | rtmax | intensity | quality | fwhm | z | source | id__0 | feature_id__0 | feature_size__0 | mz__0 | mzmin__0 | mzmax__0 | rt__0 | rtmin__0 | rtmax__0 | intensity__0 | quality__0 | fwhm__0 | z__0 | source__0 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str | int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str |
| 407 | 318 | 1 | 638.245705 | 638.245239 | 638.246704 | 0.75 m | 0.73 m | 0.77 m | 4.20e+04 | 5.97e-05 | 0.03 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 406 | 317 | 1 | 271.176873 | 271.176117 | 271.177032 | 0.82 m | 0.80 m | 0.83 m | 4.30e+04 | 6.11e-05 | 0.04 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 405 | 316 | 1 | 387.126863 | 387.126526 | 387.127106 | 1.20 m | 1.18 m | 1.23 m | 4.63e+04 | 6.57e-05 | 0.04 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 149 | 87 | 2 | 148.079803 | 148.079605 | 148.079926 | 5.43 m | 5.38 m | 5.49 m | 5.48e+04 | 1.71e-03 | 0.06 m | 1 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 404 | 315 | 1 | 779.201745 | 779.201355 | 779.202515 | 4.04 m | 3.98 m | 4.07 m | 5.68e+04 | 8.06e-05 | 0.05 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 403 | 314 | 1 | 757.558652 | 757.556519 | 757.560486 | 0.80 m | 0.76 m | 1.09 m | 5.95e+04 | 8.44e-05 | 0.04 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 402 | 313 | 1 | 750.508721 | 750.507080 | 750.510620 | 0.61 m | 0.60 m | 0.64 m | 6.23e+04 | 8.84e-05 | 0.04 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 401 | 312 | 1 | 522.203429 | 522.203064 | 522.203857 | 5.95 m | 5.93 m | 5.98 m | 6.28e+04 | 8.91e-05 | 0.03 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 120 | 68 | 3 | 597.172992 | 597.171814 | 597.178040 | 1.66 m | 1.62 m | 1.69 m | 7.23e+04 | 2.11e-03 | 0.06 m | 1 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 400 | 311 | 1 | 658.408975 | 658.407776 | 658.410034 | 0.71 m | 0.69 m | 0.76 m | 7.62e+04 | 1.08e-04 | 0.03 m | 0 | AA_sample_arabidopsis.mzML | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
The 10 most intense peaks of the Arabidopsis sample seem to be sample specific since there is not a single match with the Caulobacter sample.
Similar to run_feature_finder_metabo, MzMine2 pick_peaks features peak detection in two main processing steps ion chromatogram extraction and peak detection where different peak detection algorithms can be applied:
# mzmine2 is an extension we find in emzed name space under ext.
# We create a mopre dicrect access
mzmine = emzed.ext.mzmine2
mzmine.pick_peaks(peakmap, adap_chromatogram_builder, peak_resolver, rsp_parameters=None, verbose=False)
The peak_picker function arguments are
peakmap: PeakMap object
adap_chromatogram_builder: ADAPChromatogramBuilder object
peak_resolver: PeakDetectionBuilder object
rsp_parameters: RemoveShoulderPeaksParameters object, optional. If provided also shoulder peaks will be removed.
verbose: print more output from mzmine when verbose is True .
In contrast to openMS run_feature_finder_metabo , pick_peaks does not hand over directly feature detection parameters as function arguments, but objects defining processing step and each processing step object is configured separately. The optional rsp_parameters allows removing shoulder (satellite) peaks. Those are artificial peaks which can by generated by Orbitrap Instruments.
MzMine2 provides ADAP chromatogram extraction and peak detection algorithms Meyers et al. which we will discuss further. Let us start with ADAPChromatogramBuilder description by Meyers et al.:
Take all the data points in a datafile, sort them by their intensities, and remove those points (mostly noise) below a certain intensity threshold.
Starting with the most intense data point, the first EIC is created.
For this EIC, establish an immutable m/z range that is the data point’s where is specified by the user.
The next data point, which will be the next most intense, is added to an existing EIC if its m/z value falls within its m/z range.
If the next data point does not fall within an EICs m/z range, a new EIC is created. New EICs are only created if the point meets the minimum start intensity requirement set by the user.
An m/z range for a new EIC is created the same way as in step 3 except the boundaries will be adjusted to avoid overlapping with pre-existing EICs. As an example consider an existing EIC with m/z range (100.000,100.020) for . If the new EIC is initialized with a data point having an m/z value of 100.025, then this new EIC will have am/z range set to (100.020, 100.035) rather than (100.015, 100.035).
Repeat steps 4−6 until all the data has been processed.
Finally, a post processing step is implemented. Only EICs with a user defined number of continuous points above a user defined intensity threshold are kept.
ADAPChromatogramBuilder has 4 tuning parameters:
minimum_scan_span: Minimum number of scans over which some peak in the chromatogram must have continuous points above the noise level to be recognized as a chromatogram. The optimal value depends on the chromatography system setup. The best way to set this parameter is by studying the raw data and determining what is the typical time span of chromatographic peaks.
mz_tolerance: Maximum allowed difference between two m/z values to be considered same. The value is specified both as absolute tolerance (in m/z) and relative tolerance (in ppm). The tolerance range is calculated using maximum of the absolute and relative tolerances.
start_intensity: Points below this intensity will not be considered in starting a new chromatogram.
intensity_thresh2: This parameter is the intensity value for which intensities greater than this value can contribute to the minimum_scan_span count
Note, with start_intensity and intensity_thresh2 2 different threshold parameters are provided. It is useful to choose a higher start_intensity than intensity_thresh2 values since it allows avoiding chromatogram splits along the baseline due to noise. The parameter depends a lot on signal quality i.e. electro spray stability. openMS ff_metabo handles the same issue by allowing a user defined number of outliers (values below the intensity threshold) or alternatively, by defining a
signal frequency (not shown). All those values are instrument and sample dependent and require specific tuning. The art is to find the right balance between peak number and quality and requires some time investment. Good practice is to check for the detection of compounds known to be present in the samples at different abundances (i.e. due targeted analysis or spiked compounds).
Also note, the parameter mz_tolerance is defined by a tuple (absolute_tolerance, relative_tolerance in ppm) and the applied tolerance is defined as \(max(atol, rtol)\). Some emzed expressions have a very similar syntax i.e. column.approx_equal(other_column, atol, rtol) In contrast, all core emzed expressions use the common additive tolerance definition :math:`tol = atol + rtol *value`. To avoid unexpected results, we recommend to set one of the two tuple values to 0.
When comparing openMS run_feature_finder_metabo and MZmine2 pick_peaks performance we should configure the same or similar parameters with similar values. Let’s configure the ADAPChromatogramBuilder:
[9]:
mzmine = emzed.ext.mzmine2
chrom = mzmine.ADAPChromatogramBuilder()
chrom.intensity_thresh2 = 7e3
chrom.minimum_scan_span = 5
chrom.mz_tolerance = (0.008, 0)
chrom.start_intensity = 1e4
Next, we will define peak detection process. MzmMine 2 provides 5 different peak detectors, each with different strengths and weakness.
ADAPDetector
BaselinePeakDetector
MinimumSearchPeakDetector
NoiseAmplitudePeakDetector
SavitzkyGolayPeakDetector
In the following we will focus on the ADAPDectector since it performs best in terms of data quality.
ADAP detects peaks using continuous wavelet transformation (CWT). Such transformation simplifies peak recognition since peaks can be detected by varying wavelet scale (the principle is nicely demonstrated here. In principle, LC-MS peaks can now be detected by following along their ridges or ridgeline (max values) as function of the scaling parameter see also Meyers et al.. Configuration requires 6 parameters:
peak_duration: Range of acceptable peak lengths. Tuple (min, max) in seconds.
rt_for_cwt_scales_duration: Upper and lower bounds of retention times to be used for setting the wavelet scales. Choose a range that is similar to the range of peak widths (FWHM) in seconds expected to be found in the data.
sn_estimators: User can choose between two signal to noise estimator objects:
IntensityWindowsSNParameters was tested on LC-MS datasets and uses the peak height as the signal level and the standard deviation of intensities around the peak as the noise level
WaveletCoefficientsSNParameters was tested on GC-MS datasets and uses the continuous wavelet transform coefficients to estimate the signal and noise levels.
sn_threshold: Signal to noise ratio threshold. The minimum signal to noise ratio a peak must have to be considered a real feature. Values greater than or equal to 7 will work well and will only detect a very small number of false positive peaks.
coef_area_threshold: This is the best coefficient found by taking the inner product of the wavelet at the best scale and the peak, and then dividing by the area under the peak. Values around 100 work well for most data. Filters out bad peaks.
min_feat_height: Minimum height of a feature. The smallest intensity a peak can have and be considered a real feature. Should be the same, or similar to start_intensity value of ADAPChromatogramBuilder.
Additional Notes on rt_for_cwt_scales_duration parameter. The parameter can be interpreted as the width range of the transformed peak. The applied transformation function (mother wavelet) is the so called mexican hat, corresponding to the second derivative of the Gaussian distribution or bell curve. Most importantly, the width of the center peak corresponds to the standard deviation \(\sigma\) and is a good estimator of the chromatogram peak width FWHM range. You might also check the original configuration parameter description in the adap user manual.
Again we configure the parameters in a way to obtain results comparable with the settings we used above for run_feature_finder_metabo.
[10]:
pd = mzmine.ADAPDetector()
pd.peak_duration = (5.0, 60.0) # in seconds
pd.rt_for_cwt_scales_duration = (0.04, 3.0)
pd.sn_threshold = 10.0
pd.sn_estimators = (
mzmine.IntensityWindowsSNParameters()
) # since we have LC-MS data
pd.coef_area_threshold = 50
pd.min_feat_height = 5e4
Last not least, optional rsp_parameters allows removing shoulder (satellite) peaks. The corresponding RemoveShoulderPeaksParameters object has 2 configuration parameters:
resolution: Mass resolution is the dimensionless ratio of the mass of the peak divided by its width. Peak width is taken as the full width at half maximum intensity (FWHM). default = 100’000
peak_model: Peaks under the curve of this peak model will be removed. Allowed values: 'GAUSS', 'LORENTZ', 'LORENTZEXTENDED'; default = 'GAUSS'
Samples were acquired with a mass resolution of 30’000 at m/z 400. In case of Orbitrap instruments the resolution is
\(R = R_{ref} \sqrt(\frac{{mz}_{ref}}{mz})\)
and hence, the m/z acquisition range is important. Also keep in mind, the lower the resolution the broader the correction window width. We set it up as follows:
[11]:
rsp = mzmine.RemoveShoulderPeaksParameters(30000, "LORENTZ")
Finally, we can run mzmine feature detection:
[12]:
%%capture
ta_caulo = mzmine.pick_peaks(
peakmap_caulobacter, chrom, pd, rsp_parameters=rsp
)
[13]:
ta_caulo.sort_by("parent_id")[:10]
[13]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width |
|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType |
| 1 | - | 85.075737 | 1.31 m | 85.068855 | 85.083290 | 0.30 m | 9.83 m | 1.28e+07 | 2.01e+06 | 9.54 m |
| 2 | - | 86.059761 | 1.25 m | 86.059601 | 86.067551 | 0.59 m | 6.18 m | 1.37e+07 | 2.24e+06 | 5.59 m |
| 3 | - | 86.079033 | 1.31 m | 86.072250 | 86.082825 | 1.28 m | 8.44 m | 1.09e+06 | 8.67e+04 | 7.16 m |
| 4 | - | 86.096146 | 1.13 m | 86.096039 | 86.096184 | 1.10 m | 2.54 m | 9.79e+05 | 1.55e+05 | 1.44 m |
| 5 | - | 87.063080 | 1.25 m | 87.062820 | 87.070770 | 0.09 m | 8.53 m | 1.40e+06 | 6.27e+04 | 8.44 m |
| 6 | - | 88.050133 | 1.36 m | 88.042343 | 88.057983 | 1.07 m | 9.67 m | 1.03e+06 | 3.82e+04 | 8.60 m |
| 7 | - | 88.075447 | 1.45 m | 88.075363 | 88.075470 | 1.44 m | 1.49 m | 8.37e+05 | 4.78e+05 | 0.05 m |
| 8 | - | 88.111633 | 0.97 m | 88.103668 | 88.111786 | 0.93 m | 8.50 m | 2.16e+06 | 4.51e+04 | 7.57 m |
| 9 | - | 89.059425 | 0.74 m | 89.052620 | 89.060440 | 0.05 m | 9.98 m | 1.43e+06 | 1.65e+04 | 9.93 m |
| 10 | - | 90.054634 | 4.00 m | 90.047531 | 90.059875 | 0.58 m | 10.00 m | 1.70e+06 | 5.07e+04 | 9.42 m |
Most columns of pick_peaks and run_feature_finder_metabo output Table are the same. pick_peaks provides an additional column parent_id referring to the id of the corresponding EIC. Hence, peaks originating from the same EIC have the same parent_id value and we can sort the table by parent_id to evaluate the peak detection process:
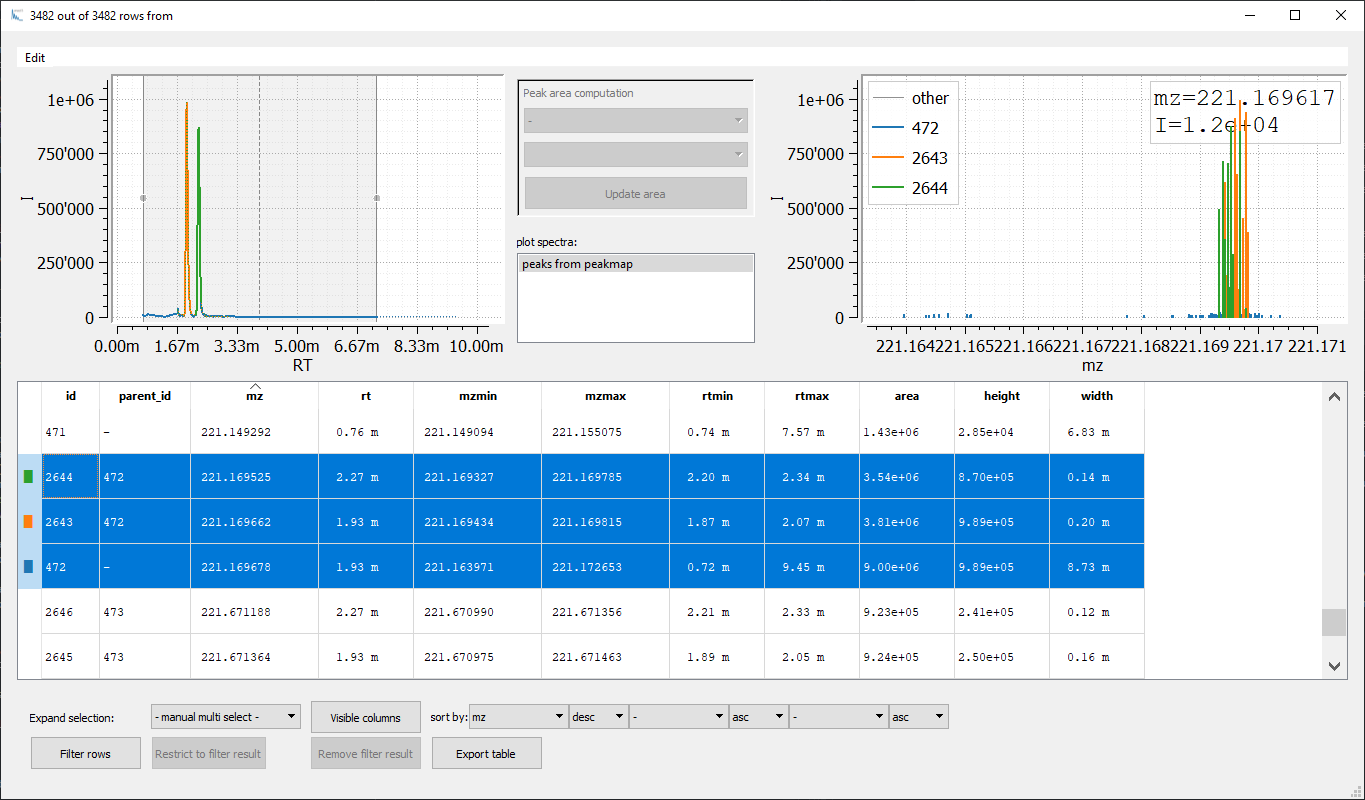
Evaluating the different chromatograms simplifies verifying peak extraction and optimization of peak detection parameters. Since we will no longer need the unsplitteded EICs for further processing, we can remove them via a simple filter command using the fact that they have an empty parent_id:
[14]:
ta_caulo = ta_caulo.filter(ta_caulo.parent_id.is_not_none())
At this state, isotopologue grouping is still missing. We can accomplish the grouping using mzmine.isotope_grouper(). To this end we have to configure the IsotopeGrouperParameters object with parameters:
mz_tolerance: Maximum allowed difference between two m/z values to be considered same. The value is specified both as absolute tolerance (in m/z) and relative tolerance (in ppm). The tolerance range is calculated using the maximum of the absolute and relative tolerances.
rt_tolerance: Maximum allowed difference between two retention time values in seconds. Defined as a tuple: (is_abs_value, value) with is_abs_value being either True or False to define if the provided value is absolute or relative. As an example we assume an RT = 100s and rtol = (True, 0.5) rt tolerance equals 0.5s and (False, 0.5) equals 50s.
monotonic_shape: If true, then monotonically decreasing height of isotope pattern is required.
maximum_charge: Maximum charge to consider for detecting the isotope patterns.
representative_isotope: peak, which should represent the whole isotope pattern. For low molecular weight compounds with monotonically decreasing isotope pattern, the most intense isotope should be representative. For high molecular weight molecules, the lowest m/z isotope may be the representative. Allowed values: 'Most intense', 'Lowest m/z'
Note, parameter mz_tolerance refers not to the mz values of isotopologues directly, but refers to the nominal isotopologue mass shift of neighboring isotopologues of an isotopologue pattern. The pattern results from the natural isotope distributions of compound elements and can be calculated (see also here). Hence, mz_tolerance should be selected in a way that it fullfills following condition for all compounds of interest:
\(\lvert (mz_{n+1, measured} -mz_{n+1, calculated}-mz_{n, measured} + mz_{n, calculated})*z\rvert\le mz_{tolerance}\)
with n corresponding to the nominal isotopologue number. Since most biomolecules are composed out of the elements C, H, N, O, P, S isotopologue shifts are mainly driven by \(^{13}C\), \(^{18}O\), \(^{34}S\) and to a certain extend by \(^{15}N\), spanning a mass range of (0.995 and 1.005), which corresponds to a mz_tolerance of about 0.005 for single charged ions. Since \(^{13}C\) is mainly repsonsible for metabolites M1 isotopologue, isotopologue grouping algorithm assums
a default isotope mass shift of 1.0033 Da corresponding to the \(^{12}C\) - \(^{13}C\) mass difference. However, high mass resolution instruments are capable to separate \(^{13}C\) and \(^{15}N\) isotopologues and if you want to include the \(^{15}N\) isotopologue peak you should increase the mass tolerance to \(\pm\) 0.008.
For most metabolites with typical elemental composition, a monotonic shape can be assumed and hence the parameter monotonic_shape should be set to True. However, keep in mind that compounds rich in S atoms or some adducts can ommit isotopologue grouping when monotopic shape is assumed, i.e. grouping of \(M_{1}\) and \(M_{2}\) of ions with a Cl adduct will fail due to the high abundance of \(^{35}Cl\).
We configure the IsotopeGrouperParameters as follows:
[15]:
ig = mzmine.IsotopeGrouperParameters()
ig.mz_tolerance = (0.005, 0)
ig.rt_tolerance = (True, 2.0) # (is_abs_value, value)
ig.monotonic_shape = True
ig.maximum_charge = 3
ig.representative_isotope = "Lowest m/z"
And we get our feature table by:
[16]:
ta_caulo_final = mzmine.isotope_grouper(ta_caulo, ig)
shutdown send alive token
got first alive token1761657723.692966
wait for first alive token
json file
/tmp/tmprzqamlhl.json
Oct 28, 2025 1:22:03 PM net.sf.mzmine.modules.rawdatamethods.rawdataimport.fileformats.MzMLReadTask run
INFO: Started parsing file /tmp/tmpf37s81n6.mzML
Oct 28, 2025 1:22:03 PM uk.ac.ebi.jmzml.MzMLElement loadProperties
WARNING: MzIdentML Configuration file: jar:file:/root/.emzed3/emzed.ext.mzmine2/3.0.0/mzmine2/MZmine-2.41.2/lib/jmzml-1.7.11.jar!/defaultMzMLElement.cfg.xml
got 1761657724.1948707
Oct 28, 2025 1:22:05 PM net.sf.mzmine.modules.rawdatamethods.rawdataimport.fileformats.MzMLReadTask run
INFO: Finished parsing /tmp/tmpf37s81n6.mzML, parsed 1066 scans
/tmp/tmpg_7cj6ax.csv
Oct 28, 2025 1:22:05 PM net.sf.mzmine.modules.peaklistmethods.isotopes.deisotoper.IsotopeGrouperTask run
INFO: Running isotopic peak grouper on
Oct 28, 2025 1:22:05 PM net.sf.mzmine.modules.peaklistmethods.isotopes.deisotoper.IsotopeGrouperTask run
INFO: Finished isotopic peak grouper on
write output file to /tmp/tmpcsbc03oz.txt
extracted 1115 deconvolved peaks
extracted 792 deconvolved peaks
!!!DONE
[17]:
ta_caulo_final.sort_by("mz", ascending=False)[:10]
[17]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int |
| 3495 | 2379 | 599.389374 | 1.01 m | 599.388489 | 599.390259 | 0.97 m | 1.08 m | 3.64e+06 | 1.41e+06 | 0.11 m | - | - | - | - |
| 3494 | 2371 | 597.875854 | 0.78 m | 597.875366 | 597.877563 | 0.74 m | 0.84 m | 3.32e+05 | 1.28e+05 | 0.09 m | 414 | 3492 | 2 | 2 |
| 3492 | 2370 | 597.374146 | 0.78 m | 597.373413 | 597.375305 | 0.73 m | 0.83 m | 5.35e+05 | 1.74e+05 | 0.09 m | 414 | 3492 | 2 | 2 |
| 3493 | 2370 | 597.374146 | 0.54 m | 597.373596 | 597.376770 | 0.53 m | 0.61 m | 2.21e+05 | 7.46e+04 | 0.07 m | - | - | - | - |
| 3491 | 2368 | 596.317200 | 0.50 m | 596.312622 | 596.318970 | 0.47 m | 0.54 m | 4.72e+05 | 2.11e+05 | 0.07 m | 40 | 3489 | 3 | 1 |
| 3490 | 2367 | 595.315369 | 0.48 m | 595.314880 | 595.316895 | 0.47 m | 0.56 m | 2.46e+06 | 1.19e+06 | 0.10 m | 40 | 3489 | 3 | 1 |
| 3489 | 2365 | 594.312195 | 0.48 m | 594.310303 | 594.312561 | 0.47 m | 0.56 m | 8.60e+06 | 4.07e+06 | 0.10 m | 40 | 3489 | 3 | 1 |
| 3486 | 2351 | 591.876099 | 0.72 m | 591.875061 | 591.877075 | 0.67 m | 0.78 m | 3.99e+05 | 1.83e+05 | 0.10 m | 331 | 3484 | 4 | 2 |
| 3487 | 2351 | 591.875244 | 1.15 m | 591.873657 | 591.876282 | 1.12 m | 1.24 m | 2.62e+05 | 6.80e+04 | 0.12 m | 331 | 3484 | 4 | 2 |
| 3488 | 2351 | 591.875092 | 0.96 m | 591.873291 | 591.877380 | 0.91 m | 0.98 m | 1.56e+05 | 6.00e+04 | 0.07 m | 331 | 3484 | 4 | 2 |
Note, only grouped peaks obtain additional identifiers, hence columns isotope_group_id and isotope_base_peak contain None values if peaks were not grouped.
We can use the emzed Table method join to compare the feature detection results. Ideally, both methods will result in the same mz and rt values. We can test this using Table.join allowing only small mz and rt tolerances:
[18]:
adap = ta_caulo_final
ff = t_caulo
mztol = (0.001, 0.0)
rttol = (2.0, 0.0) # in the range of about one spectrum distance
comp_adap_ff = adap.join(
ff,
adap.mz.approx_equal(ff.mz, *mztol)
& adap.rt.approx_equal(ff.rt, *rttol),
)
print("number of common peaks:", len(comp_adap_ff))
shutdown send alive token
number of common peaks: 736
Note, the length of table comp_adap_ff does not necessarily correspond to the number of common peaks since in principle, a peak in one table can match several peaks in the other table and vice versa, leading to multiple entries for each peak. Here, a more general way to determine the number of common peaks:
[19]:
ids = comp_adap_ff.id.to_list()
no_peaks = len(set(ids))
print("number of common peaks:", no_peaks)
number of common peaks: 736
Ideally, both feature detection approaches result identical m/z and rt values for the same peaks. We can add a column to show the mz differences:
[20]:
t = comp_adap_ff
t.add_or_replace_column(
"mz_delta", t.mz - t.mz__0, emzed.MzType, format_="%.1e"
)
t.add_or_replace_column(
"mz_delta",
t.apply(abs, t.mz_delta),
emzed.MzType,
format_="%.1e",
)
t = t.sort_by("mz_delta", ascending=False)
t[:10]
[20]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge | id__0 | feature_id__0 | feature_size__0 | mz__0 | mzmin__0 | mzmax__0 | rt__0 | rtmin__0 | rtmax__0 | intensity__0 | quality__0 | fwhm__0 | z__0 | source__0 | mz_delta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int | int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str | MzType |
| 2718 | 618 | 245.605164 | 1.51 m | 245.603516 | 245.610641 | 1.49 m | 1.56 m | 1.67e+05 | 6.15e+04 | 0.07 m | - | - | - | - | 575 | 341 | 2 | 245.606116 | 245.602371 | 245.610641 | 1.53 m | 1.49 m | 1.58 m | 2.02e+05 | 1.67e-04 | 0.06 m | 2 | AA_sample_caulobacter.mzML | 9.5e-04 |
| 3187 | 1661 | 447.270050 | 0.53 m | 447.269714 | 447.270508 | 0.52 m | 0.59 m | 9.05e+05 | 3.96e+05 | 0.07 m | - | - | - | - | 581 | 346 | 1 | 447.269209 | 447.259308 | 447.272034 | 0.53 m | 0.47 m | 0.66 m | 7.04e+05 | 1.64e-04 | 0.03 m | 0 | AA_sample_caulobacter.mzML | 8.4e-04 |
| 3338 | 2033 | 520.332031 | 0.74 m | 520.330017 | 520.333252 | 0.70 m | 0.79 m | 6.78e+05 | 2.83e+05 | 0.09 m | - | - | - | - | 660 | 412 | 1 | 520.332712 | 520.330017 | 520.344360 | 0.74 m | 0.71 m | 0.82 m | 5.35e+05 | 1.25e-04 | 0.03 m | 0 | AA_sample_caulobacter.mzML | 6.8e-04 |
| 3422 | 2236 | 564.327545 | 0.52 m | 564.321594 | 564.328308 | 0.50 m | 0.57 m | 1.33e+06 | 5.86e+05 | 0.07 m | - | - | - | - | 390 | 195 | 2 | 564.328171 | 564.321594 | 564.341370 | 0.52 m | 0.50 m | 0.64 m | 1.17e+06 | 3.53e-04 | 0.05 m | 1 | AA_sample_caulobacter.mzML | 6.3e-04 |
| 2675 | 534 | 230.838531 | 1.53 m | 230.835220 | 230.840469 | 1.46 m | 1.60 m | 3.98e+06 | 9.45e+05 | 0.14 m | 132 | 2675 | 2 | 3 | 244 | 110 | 1 | 230.837950 | 230.835220 | 230.840469 | 1.53 m | 1.45 m | 1.59 m | 3.26e+06 | 7.61e-04 | 0.08 m | 0 | AA_sample_caulobacter.mzML | 5.8e-04 |
| 3491 | 2368 | 596.317200 | 0.50 m | 596.312622 | 596.318970 | 0.47 m | 0.54 m | 4.72e+05 | 2.11e+05 | 0.07 m | 40 | 3489 | 3 | 1 | 113 | 45 | 3 | 596.317778 | 596.312622 | 596.320618 | 0.49 m | 0.47 m | 0.56 m | 4.03e+05 | 2.15e-03 | 0.05 m | 1 | AA_sample_caulobacter.mzML | 5.8e-04 |
| 3262 | 1853 | 484.314926 | 1.57 m | 484.312286 | 484.315338 | 1.54 m | 1.64 m | 4.49e+05 | 1.68e+05 | 0.10 m | 123 | 3257 | 6 | 3 | 332 | 155 | 3 | 484.314352 | 484.312286 | 484.315338 | 1.58 m | 1.50 m | 1.63 m | 4.71e+05 | 5.00e-04 | 0.06 m | 3 | AA_sample_caulobacter.mzML | 5.7e-04 |
| 3375 | 2097 | 533.306763 | 0.52 m | 533.304932 | 533.307312 | 0.50 m | 0.59 m | 1.41e+06 | 6.08e+05 | 0.09 m | 181 | 3375 | 2 | 1 | 395 | 199 | 2 | 533.306200 | 533.296936 | 533.307312 | 0.52 m | 0.46 m | 0.61 m | 1.19e+06 | 3.38e-04 | 0.04 m | 1 | AA_sample_caulobacter.mzML | 5.6e-04 |
| 3293 | 1933 | 499.283325 | 1.55 m | 499.281006 | 499.284668 | 1.48 m | 1.58 m | 6.57e+05 | 1.74e+05 | 0.10 m | 23 | 3286 | 3 | 1 | 41 | 15 | 3 | 499.282783 | 499.279083 | 499.284668 | 1.54 m | 1.47 m | 1.58 m | 6.34e+05 | 5.55e-03 | 0.06 m | 1 | AA_sample_caulobacter.mzML | 5.4e-04 |
| 3434 | 2261 | 570.339050 | 0.80 m | 570.331909 | 570.341797 | 0.75 m | 0.84 m | 3.59e+05 | 1.34e+05 | 0.09 m | 291 | 3431 | 3 | 3 | 535 | 308 | 2 | 570.338545 | 570.331909 | 570.341797 | 0.80 m | 0.75 m | 0.86 m | 2.92e+05 | 1.96e-04 | 0.05 m | 3 | AA_sample_caulobacter.mzML | 5.1e-04 |
We observe rather small m/z variances at the 4th digit which are below mass accuracy of the instrument. Exceptions seem to be due to bad quality i.e. peak id==2722 turned out to be a satellite peak which has not been removed. Summarized, peaks detected with both methods give the same results for common detected peaks.
Next we evaluate peaks exclusively extracted with only one out of the two detectors: the joined table provides peaks ids of those peaks detected with both tools. With the method left_join we can also find peaks detected with only one out of the two tools, since rows of the joined table contain only None values.
[21]:
# mzmine2
comp_adap_ff = adap.left_join(
ff,
adap.mz.approx_equal(ff.mz, *mztol)
& adap.rt.approx_equal(ff.rt, *rttol),
)
adap_only = comp_adap_ff.filter(comp_adap_ff.id__0.is_none())
# open ms
comp_ff_adap = ff.left_join(
adap,
ff.mz.approx_equal(adap.mz, *mztol)
& ff.rt.approx_equal(adap.rt, *rttol),
)
ff_only = comp_ff_adap.filter(comp_ff_adap.id__0.is_none())
print(f"only by ADAP: {len(adap_only)}")
print(f"only by run_feature_finder_metabo: {len(ff_only)}")
only by ADAP: 379
only by run_feature_finder_metabo: 308
Note, here we do not build a set from id values to count the number of peaks, since by definition there are exists no similar peak in the joint table.
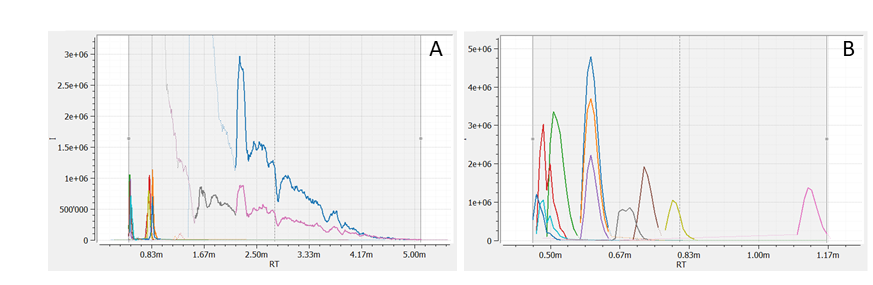
Figure 5: Top 10 peaks exclusively detected with ff_metabo (A) and ADAP (B).
When evaluating exclusive peaks, both approaches miss significant peaks (Figure 5). For given parameters, ff_metabo detects about 4 % more peaks than ADAP but it ff_metabo misses more high quality peaks.
SUMMARY: Both approaches give similar results with a slightly better performance of the ADAP. However, some further parameter optimization might close the gap between the two methods. ADAP has a clear advantage in case of Orbitrap instruments data since it can directly remove shoulder peaks. On the other hand ff_metabo is about 100x faster than ADAP. This is a clear advantage when analyzing huge data sets.
emzed.adducts module largely enhances adduct annotation. Note, that here we do not use the term adduct properly, since we also consider protonation and deprotonation “adducts”. First, it provides detailed information about all common adducts. In total 54 adducts in positive and negative ionization mode are provided. To get detailed information about all available adducts in both modes use the command
[22]:
emzed.adducts.all[:10]
[22]:
| id | adduct_name | m_multiplier | adduct_add | adduct_sub | z | sign_z |
|---|---|---|---|---|---|---|
| int | str | int | str | str | int | int |
| 0 | M-3H | 1 | H3 | 3 | -1 | |
| 1 | M-2H | 1 | H2 | 2 | -1 | |
| 2 | M- | 1 | 1 | -1 | ||
| 3 | M-H | 1 | H | 1 | -1 | |
| 4 | M-H2O-H | 1 | H2OH | 1 | -1 | |
| 5 | M+Na-2H | 1 | Na | H2 | 1 | -1 |
| 6 | M+Cl | 1 | Cl | 1 | -1 | |
| 7 | M+K-2H | 1 | K | H2 | 1 | -1 |
| 8 | M+KCl-H | 1 | KCl | H | 1 | -1 |
| 9 | M+FA-H | 1 | H2CO2 | H | 1 | -1 |
[23]:
emzed.adducts.all[-10:]
[23]:
| id | adduct_name | m_multiplier | adduct_add | adduct_sub | z | sign_z |
|---|---|---|---|---|---|---|
| int | str | int | str | str | int | int |
| 44 | M+3ACN+2H | 1 | (C2H3N)3H2 | 2 | 1 | |
| 45 | M+ACN+2H | 1 | (C2H3N)1H2 | 2 | 1 | |
| 46 | M+2H | 1 | H2 | 2 | 1 | |
| 47 | M+H+Na | 1 | HNa | 2 | 1 | |
| 48 | M+H+K | 1 | HK | 2 | 1 | |
| 49 | M+2Na | 1 | Na2 | 2 | 1 | |
| 50 | M+3H | 1 | H3 | 3 | 1 | |
| 51 | M+2H+Na | 1 | (H2)1Na | 3 | 1 | |
| 52 | M+3Na | 1 | Na3 | 3 | 1 | |
| 53 | M+2Na+H | 1 | (Na2)1H | 3 | 1 |
For space reasons we only list the first 10 and the last 10 adducts for each mode. Multitude of adducts subsets are also directly available i.e.
[24]:
adds = emzed.adducts.positive_single_charged
adds
[24]:
| id | adduct_name | m_multiplier | adduct_add | adduct_sub | z | sign_z |
|---|---|---|---|---|---|---|
| int | str | int | str | str | int | int |
| 19 | M+ | 1 | 1 | 1 | ||
| 20 | M+H | 1 | H | 1 | 1 | |
| 21 | M+NH4 | 1 | NH4 | 1 | 1 | |
| 22 | M+Na | 1 | Na | 1 | 1 | |
| 23 | M+H-2H2O | 1 | H | (H2O)2 | 1 | 1 |
| 24 | M+H-H2O | 1 | H | H2O | 1 | 1 |
| 25 | M+K | 1 | K | 1 | 1 | |
| 26 | M+ACN+H | 1 | C2H3NH | 1 | 1 | |
| 27 | M+2ACN+H | 1 | (C2H3N)2H | 1 | 1 | |
| 28 | M+ACN+Na | 1 | (C2H3N)1Na | 1 | 1 | |
| 29 | M+2Na-H | 1 | Na2 | H | 1 | 1 |
| 30 | M+Li | 1 | Li | 1 | 1 | |
| 31 | M+CH3OH+H | 1 | CH3OHH | 1 | 1 | |
| 32 | M+2K-H | 1 | K2 | H | 1 | 1 |
| 33 | M+IsoProp+H | 1 | (C3H8O)1H | 1 | 1 | |
| 34 | M+IsoProp+Na+H | 1 | (C3H8O)1NaH | 1 | 1 | |
| 35 | M+DMSO+H | 1 | (C2H6OS)1H | 1 | 1 | |
| 36 | 2M+H | 2 | H | 1 | 1 | |
| 37 | 2M+NH4 | 2 | NH4 | 1 | 1 | |
| 38 | 2M+Na | 2 | Na | 1 | 1 | |
| 39 | 2M+K | 2 | K | 1 | 1 | |
| 40 | 2M+ACN+H | 2 | (C2H3N)1H | 1 | 1 | |
| 41 | 2M+ACN+Na | 2 | (C2H3N)1Na | 1 | 1 |
Columns m_multiplier, adduct_add, adduct_sub and z are required to calculate the correct neutral mass. Provided adduct tables can be directly used for adduct asignment with method:
[25]:
annotate = emzed.annotate.annotate_adducts
help(annotate) # more explanat
Help on function annotate_adducts in module emzed.annotate.annotate_adducts:
annotate_adducts(peaks, adducts, mz_tol, rt_tol, explained_abundance=0.2)
attempts to group peaks as adducts.
required column names are `mz` and `rt` only. all other columns will be ignored.
Note that you should select only those adducts for assignment which are likely to be present. Some adducts i.e. acetate are more difficult to interprete since the mass shift can also origin from an in source fragmentation of i.e. a sugar moiety. We will select most common adducts from our adducts table by columnn id:
[26]:
%%capture
ids = [20, 21, 22, 24, 25, 32, 36, 38, 39]
adducts = adds.filter(adds.id.is_in(ids))
t_annotate = annotate(adap, adducts, 0.002, 1.5, 0.3)
[27]:
t_annotate.sort_by("adduct_cluster_id", ascending=False)[:10]
[27]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge | adduct_name | adduct_isotopes | adduct_isotopes_abundance | adduct_m0 | adduct_cluster_id |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int | str | str | float | MzType | int |
| 2460 | 123 | 143.154099 | 6.24 m | 143.154053 | 143.154343 | 6.20 m | 6.29 m | 2.51e+05 | 6.59e+04 | 0.09 m | - | - | - | - | M+H | +[1]H | 0.999885 | 142.146822 | 148 |
| 2506 | 197 | 160.180588 | 6.22 m | 160.180466 | 160.180832 | 6.16 m | 6.40 m | 1.26e+08 | 2.56e+07 | 0.24 m | 5 | 2506 | 3 | 1 | M+NH4 | +[1]H4 [14]N | 0.995862 | 142.146762 | 148 |
| 2475 | 141 | 151.035149 | 6.10 m | 151.034973 | 151.035324 | 6.08 m | 6.19 m | 9.79e+06 | 2.55e+06 | 0.10 m | 67 | 2475 | 2 | 1 | M+NH4 | +[1]H4 [14]N | 0.995862 | 133.001323 | 147 |
| 2771 | 718 | 267.013214 | 6.10 m | 267.012726 | 267.013519 | 6.08 m | 6.19 m | 6.60e+05 | 1.85e+05 | 0.10 m | - | - | - | - | 2M+H | +[1]H | 0.999885 | 133.002969 | 147 |
| 2391 | 15 | 94.044815 | 6.11 m | 94.044685 | 94.044945 | 6.09 m | 6.18 m | 3.34e+05 | 1.16e+05 | 0.09 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 76.010990 | 146 |
| 2481 | 150 | 153.032631 | 6.10 m | 153.032425 | 153.032776 | 6.08 m | 6.19 m | 1.40e+06 | 3.69e+05 | 0.10 m | - | - | - | - | 2M+H | +[1]H | 0.999885 | 76.012677 | 146 |
| 2683 | 555 | 237.090317 | 6.01 m | 237.090012 | 237.091080 | 5.98 m | 6.09 m | 1.23e+06 | 5.54e+05 | 0.11 m | - | - | - | - | 2M+H | +[1]H | 0.999885 | 118.041520 | 145 |
| 2751 | 681 | 259.072266 | 6.01 m | 259.071991 | 259.072571 | 6.00 m | 6.07 m | 1.47e+05 | 7.63e+04 | 0.08 m | - | - | - | - | 2M+Na | +[23]Na | 1.000000 | 118.041522 | 145 |
| 2683 | 555 | 237.090317 | 6.01 m | 237.090012 | 237.091080 | 5.98 m | 6.09 m | 1.23e+06 | 5.54e+05 | 0.11 m | - | - | - | - | M+H | +[1]H | 0.999885 | 236.083040 | 144 |
| 2751 | 681 | 259.072266 | 6.01 m | 259.071991 | 259.072571 | 6.00 m | 6.07 m | 1.47e+05 | 7.63e+04 | 0.08 m | - | - | - | - | M+Na | +[23]Na | 1.000000 | 236.083045 | 144 |
annotate_adducts adds colums adduct_name, adduct_isotopes, adduct_abundance, adduct_m0 and adduct_cluster_id. It provides detailed information about the grouping process and manual evaluation is straightforward. The more adducts you add the more groupings are possible and adducts might be present in more than 1 adduct group, which can be deduced from table length:
[28]:
print("before:", len(adap), "\nafter :", len(t_annotate))
before: 1115
after : 311
With a few lines of code we can extract those cases:
[29]:
from collections import Counter
id2counts = Counter(t_annotate.id)
# peaks with ids present more than ones show ambiguous grouping
ids = [key for key, counts in id2counts.items() if counts > 1]
# next we can filter our table for selected peaks
t = t_annotate.filter(t_annotate.id.is_in(ids))
# we now obtain the associated adduct_cluster_ids
ac_is = t.adduct_cluster_id.to_list()
# now we can access the subtable of interest:
t_cases = t_annotate.filter(t_annotate.adduct_cluster_id.is_in(ac_is))
t_cases[:10]
[29]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge | adduct_name | adduct_isotopes | adduct_isotopes_abundance | adduct_m0 | adduct_cluster_id |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int | str | str | float | MzType | int |
| 2382 | 2 | 86.059761 | 1.25 m | 86.059662 | 86.059853 | 1.22 m | 1.35 m | 6.63e+06 | 2.24e+06 | 0.13 m | 73 | 2382 | 3 | 1 | M+H | +[1]H | 0.999885 | 85.052485 | 0 |
| 2407 | 39 | 112.050262 | 1.63 m | 112.050102 | 112.050461 | 1.59 m | 1.72 m | 8.53e+05 | 2.85e+05 | 0.13 m | - | - | - | - | M+H | +[1]H | 0.999885 | 111.042986 | 2 |
| 2407 | 39 | 112.050262 | 1.63 m | 112.050102 | 112.050461 | 1.59 m | 1.72 m | 8.53e+05 | 2.85e+05 | 0.13 m | - | - | - | - | 2M+H | +[1]H | 0.999885 | 55.521493 | 3 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 |
| 2424 | 67 | 122.924286 | 2.16 m | 122.924202 | 122.924393 | 2.10 m | 2.23 m | 3.22e+06 | 9.15e+05 | 0.12 m | - | - | - | - | M+K | +[39]K | 0.932581 | 83.961128 | 99 |
| 2444 | 97 | 134.032135 | 1.63 m | 134.031860 | 134.032349 | 1.60 m | 1.68 m | 2.37e+05 | 1.08e+05 | 0.08 m | - | - | - | - | M+Na | +[23]Na | 1.000000 | 111.042914 | 2 |
| 2444 | 97 | 134.032135 | 1.63 m | 134.031860 | 134.032349 | 1.60 m | 1.68 m | 2.37e+05 | 1.08e+05 | 0.08 m | - | - | - | - | 2M+Na | +[23]Na | 1.000000 | 55.521457 | 3 |
| 2479 | 145 | 152.056465 | 1.92 m | 152.056320 | 152.056839 | 1.88 m | 2.05 m | 7.97e+05 | 2.44e+05 | 0.17 m | 346 | 2479 | 2 | 1 | M+H | +[1]H | 0.999885 | 151.049189 | 90 |
| 2479 | 145 | 152.056465 | 1.92 m | 152.056320 | 152.056839 | 1.88 m | 2.05 m | 7.97e+05 | 2.44e+05 | 0.17 m | 346 | 2479 | 2 | 1 | 2M+H | +[1]H | 0.999885 | 75.524594 | 91 |
| 2484 | 153 | 153.102066 | 1.25 m | 153.101929 | 153.102234 | 1.19 m | 1.42 m | 2.00e+07 | 5.76e+06 | 0.23 m | - | - | - | - | M+H-H2O | +[1]H -[1]H2 [16]O | 0.997226 | 170.105355 | 6 |
When scrolling to the right end of the resulting table we observe that multiple grouping was due to selecting the adducts M+X and 2M+X. If both adducts result identical mass shifts but different monoisotopic masses m0, the same peaks are grouped twice but with different adduct_custer_id. Hence, selecting M+X and 2M+X at the same time must result in multiple grouping. Choose possible adducts with care!
MzMine2 supports adduct annotation, thereby correctly using the term adduct, meaning protonated and deprotonated ions are not included. The adduct search algorithm only annotates potential adducts. However, since a grouping id is missing it is hardly possible to evaluate the quality of the grouping process. MzMine2 search_adducts is only suited for adduct peak removing. Although grouping of molecule ions of different charge state is possible with Mzmine2, the required method Peak
complex search is currently not supported by emzed and therefore, we recommend using the emzed.annotate_adducts() method described above.
How to group adducts with the MzMine2 extension? The function adduct_search(peaks, parameters) with attribute peaks being a peaks table with required columns id, mz, rt, height and wih attribute parameters being an mzmine AdductSearchParameters(rt_tolerance, adducts, mz_tolerance, max_adduct_height) object. The default parameters are configured as follows:
[30]:
rt_tolerance = (False, 1.0)
adducts = (
("[M+Na-H]", 21.9825),
("[M+K-H]", 37.9559),
("[M+Mg-2H]", 21.9694),
("[M+NH3]", 17.0265),
("[M+H3PO4]", 97.9769),
("[M+H2SO4]", 97.9674),
("[M+H2CO3]", 62.0004),
("[(Deuterium)]glycerol", 5.0),
)
mz_tolerance = (0.001, 0)
max_adduct_height = 1.0
rt_tolerance: a tuple (is_abolute_value, \(\Delta RT\)) that allows defining absolute difference by is_abolute_value (True, False). By default relative rt value differences are used. However, since we group coeluting peak the allowed tolerance is rather depending on the peak width (shape) than on the RT itself and absolute values should be used.
adducts: are tuples (adduct name, mass difference to molecular ion) and any single charged adduct can be defined. Note: Default values contain not the adduct ion itself but the mass shift relative to the main ion (normally M+H, M-H). Morevoer mass shifts of both ionization modes are listed and hence the list must be adapted to obtain meainingful results.
mz_tolerance: a tuple (absolute, relative) with absolute and relative allowed maximal adduct mass difference. Remember, if you provide both, a relative and an absolute difference, the higher of both values is applied.
max_adduct_height: Maximum height of the recognized adduct peak, relative to the main peak. By default peaks of the same height are allowed.
Again, we can use emzed.adducts module to build our adducts search list
[31]:
adds = emzed.adducts.positive_single_charged
ids = [21, 22, 24, 25, 32]
adducts = adds.filter(adds.id.is_in(ids))
a = adducts
# we calculate the mass shift relative to the M+H ion
expr = (
a.apply(emzed.mass.of, a.adduct_add)
- a.apply(emzed.mass.of, a.adduct_sub)
- emzed.mass.H1
)
a.add_column("mass_shift", expr, float)
# We build the (name, mass_shift) tuple with mass shift relative to M+H
adducts = list(zip(a.adduct_name, a.mass_shift))
adducts
[31]:
[('M+NH4', 17.0265490957),
('M+Na', 21.981944248999998),
('M+H-H2O', -18.0105650638),
('M+K', 37.955881648100004),
('M+2K-H', 75.91176329620001)]
We can now setup the AdductSearchParameter object:
params = mzmine.AdductSearchParameters()
params.adducts = adducts
params.mz_tolerance=(0.001, 0.0)
params.rt_tolerance=(True, 2.0)
params_max_adduct_height=1.0
and we can run adduct_search:
ta_caulo_annotated = mzmine.adduct_search(t_caulo, params)
[32]:
annotated = ta_caulo_annotated.filter(
ta_caulo_annotated.adduct_annotation.is_not_none()
)
annotated[:10]
[32]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge | adduct_annotation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int | str |
| 2444 | 97 | 134.032135 | 1.63 m | 134.031860 | 134.032349 | 1.60 m | 1.68 m | 2.37e+05 | 1.08e+05 | 0.08 m | - | - | - | - | M+Na 21.9819 m/z adduct of 112.0503 m/z |
| 2484 | 153 | 153.102066 | 1.25 m | 153.101929 | 153.102234 | 1.19 m | 1.42 m | 2.00e+07 | 5.76e+06 | 0.23 m | - | - | - | - | M+H-H2O -18.0106 m/z adduct of 171.1126 m/z |
| 2488 | 159 | 154.086075 | 0.55 m | 154.085953 | 154.086288 | 0.52 m | 0.61 m | 3.39e+05 | 1.01e+05 | 0.10 m | - | - | - | - | M+H-H2O -18.0106 m/z adduct of 172.0966 m/z |
| 2490 | 160 | 154.105362 | 1.25 m | 154.105255 | 154.105515 | 1.18 m | 1.33 m | 1.38e+06 | 4.84e+05 | 0.15 m | 2 | 2483 | 4 | 1 | M+H-H2O -18.0106 m/z adduct of 172.1159 m/z |
| 2534 | 253 | 174.038376 | 1.92 m | 174.038132 | 174.038559 | 1.89 m | 2.03 m | 8.39e+05 | 2.43e+05 | 0.14 m | - | - | - | - | M+Na 21.9819 m/z adduct of 152.0565 m/z |
| 2562 | 305 | 186.038345 | 1.32 m | 186.038132 | 186.038528 | 1.30 m | 1.38 m | 1.38e+05 | 5.29e+04 | 0.08 m | - | - | - | - | M+Na 21.9819 m/z adduct of 164.0565 m/z |
| 2745 | 681 | 259.072266 | 6.01 m | 259.071991 | 259.072571 | 6.00 m | 6.07 m | 1.47e+05 | 7.63e+04 | 0.08 m | - | - | - | - | M+Na 21.9819 m/z adduct of 237.0903 m/z |
| 2751 | 691 | 261.160095 | 0.52 m | 261.156097 | 261.160767 | 0.51 m | 0.59 m | 2.15e+05 | 7.31e+04 | 0.08 m | - | - | - | - | M+H-H2O -18.0106 m/z adduct of 279.1706 m/z |
| 2766 | 720 | 267.130951 | 1.53 m | 267.130585 | 267.131561 | 1.45 m | 1.56 m | 1.05e+06 | 3.37e+05 | 0.11 m | - | - | - | - | M+Na 21.9819 m/z adduct of 245.1494 m/z |
| 2787 | 778 | 276.070312 | 3.98 m | 276.069855 | 276.070496 | 3.95 m | 4.04 m | 1.80e+05 | 6.68e+04 | 0.10 m | - | - | - | - | M+Na 21.9819 m/z adduct of 254.0883 m/z |
The emzed.db module supports database matching using a local subset of the pubchem data base, which comprises all compounds annotated in KEGG, BIOCYC, and HMDB. When using the local database for the first time, you have to run the command:
[33]:
emzed.db.update_pubchem()
update pubchem: 00:00:16.6 total runtime
got 243111 entries from pubchem_2025-10-02_04-03.gz
you can access emzed.db.pubchem now
Downloading the database might take several minutes. Once the download is finished you can access the pubchem database as Table with the command:
[34]:
pc = emzed.db.pubchem
pc[:5]
[34]:
| cid | mw | m0 | mf | iupac | synonyms | inchi | inchikey | smiles | is_in_kegg | is_in_hmdb | is_in_biocyc | url |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str | float | float | str | str | str | str | str | str | bool | bool | bool | str |
| 176538272 | 453.4000 | 453.105985 | C23H19NO9 | [(1R,2R,3R,4S)-1-acetyloxy-2,4,9-trihydroxy-11-(iminomethylidene)-2-methyl-5,10-dioxo-3,4-dihydro-1H-benzo[b]fluoren-3-yl] acetate | InChI=1S/C23H19NO9/c1-8(25)32-21-15-11(7-24)14-16(17(15)20(30)22(23(21,3)31)33-9(2)26)18(28)10-5-4-6-12(27)13(10)19(14)29/h4-6,20-22,24,27,30-31H,1-3H3/t20-,21+,22+,23+/m0/s1 | NXQFHTZENLUMIP-WBADGQHESA-N | CC(=O)OC1C(C2=C(C(C1(C)O)OC(=O)C)C(=C=N)C3=C2C(=O)C4=C(C3=O)C(=CC=C4)O)O | False | False | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176538272 | |
| 176528301 | 226.2800 | 226.165443 | C8H18N8 | 1-[(E)-[(4E)-4-(carbamimidoylhydrazinylidene)hexan-3-ylidene]amino]guanidine | InChI=1S/C8H18N8/c1-3-5(13-15-7(9)10)6(4-2)14-16-8(11)12/h3-4H2,1-2H3,(H4,9,10,15)(H4,11,12,16)/b13-5+,14-6+ | HXWLLFLQUZOBFF-ACFHMISVSA-N | CCC(=NNC(=N)N)C(=NNC(=N)N)CC | False | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176528301 | |
| 176527043 | 239.2300 | 239.101840 | C9H13N5O3 | (10R,11R)-10-hydroxy-4-imino-11-methyl-12-oxa-3,5,7,13-tetrazatricyclo[7.3.1.01,6]tridec-6-en-2-one | O2'-4a-cyclic-tetrahydrobiopterin, 4a-Cyc-BH(,4), 4a-Cyclic-tetrahydrobiopterin, (10R,11R)-4-amino-10-hydroxy-11-methyl-12-oxa-3,5,7,13-tetraazatricyclo[7.3.1.0^{1,6}]trideca-3,5-dien-2-one | InChI=1S/C9H13N5O3/c1-3-5(15)4-2-11-6-9(14-4,17-3)7(16)13-8(10)12-6/h3-5,14-15H,2H2,1H3,(H3,10,11,12,13,16)/t3-,4?,5+,9?/m1/s1 | JZTDUNXDDUJXRZ-UPYOWCHBSA-N | CC1C(C2CN=C3C(N2)(O1)C(=O)NC(=N)N3)O | False | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176527043 |
| 176522594 | 583.6000 | 583.281324 | C21H41N7O12 | 1-[5-(diaminomethylideneamino)-2-[3-[4,5-dihydroxy-6-(hydroxymethyl)-3-(methylamino)oxan-2-yl]oxy-4-hydroxy-4-(hydroxymethyl)-5-methyloxolan-2-yl]oxy-3,4,6-trihydroxycyclohexyl]guanidine | DTXSID60859253, N,N'-[4-({5-Deoxy-2-O-[2-deoxy-2-(methylamino)hexopyranosyl]-3-C-(hydroxymethyl)pentofuranosyl}oxy)-2,5,6-trihydroxycyclohexane-1,3-diyl]diguanidine | InChI=1S/C21H41N7O12/c1-5-21(36,4-30)16(40-17-9(26-2)13(34)10(31)6(3-29)38-17)18(37-5)39-15-8(28-20(24)25)11(32)7(27-19(22)23)12(33)14(15)35/h5-18,26,29-36H,3-4H2,1-2H3,(H4,22,23,27)(H4,24,25,28) | ASXBYYWOLISCLQ-UHFFFAOYSA-N | CC1C(C(C(O1)OC2C(C(C(C(C2O)O)N=C(N)N)O)NC(=N)N)OC3C(C(C(C(O3)CO)O)O)NC)(CO)O | False | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176522594 |
| 131802965 | 197.2300 | 89.105194 | C10H15NO3 | 2-(1-oxoocta-1,7-dien-2-ylamino)acetic acid | InChI=1S/C10H15NO3/c1-2-3-4-5-6-9(8-12)11-7-10(13)14/h2,11H,1,3-7H2,(H,13,14) | OEFQVBVKPTYRNN-UHFFFAOYSA-N | C=CCCCCC(=C=O)NCC(=O)O | False | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=131802965 |
Colummns is_kegg, is_hmdb, and is_biocyc allows further constraining the database to a single source. To obtain the KEGG database one can apply the Table filter method:
[35]:
kegg = pc.filter(pc.is_in_kegg)
kegg[:5]
[35]:
| cid | mw | m0 | mf | iupac | synonyms | inchi | inchikey | smiles | is_in_kegg | is_in_hmdb | is_in_biocyc | url |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str | float | float | str | str | str | str | str | str | bool | bool | bool | str |
| 176516537 | 193.6800 | 193.109423 | C6H16ClN5 | 2-butyl-1-carbamimidoylguanidine;hydrochloride | Buformin hydrochloride (JP18), D02206 | InChI=1S/C6H15N5.ClH/c1-2-3-4-10-6(9)11-5(7)8;/h2-4H2,1H3,(H6,7,8,9,10,11);1H | KKLWSPPIRBIEOV-UHFFFAOYSA-N | CCCCN=C(N)NC(=N)N.Cl | True | False | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176516537 |
| 176490458 | 607.4000 | 607.140860 | C22H29F2N5O9P2 | 2-amino-1-[3-(4-fluoroanilino)-2-(4-fluorophenyl)-8,8-dimethyl-5,6-dihydroimidazo[1,2-a]pyrazin-7-yl]ethanone;phosphoric acid | Ganaplacide phosphate (USAN), D13145, 3051901-82-4 | InChI=1S/C22H23F2N5O.2H3O4P/c1-22(2)21-27-19(14-3-5-15(23)6-4-14)20(26-17-9-7-16(24)8-10-17)28(21)11-12-29(22)18(30)13-25;2*1-5(2,3)4/h3-10,26H,11-13,25H2,1-2H3;2*(H3,1,2,3,4) | JWLUWAGOLQWKBS-UHFFFAOYSA-N | CC1(C2=NC(=C(N2CCN1C(=O)CN)NC3=CC=C(C=C3)F)C4=CC=C(C=C4)F)C.OP(=O)(O)O.OP(=O)(O)O | True | False | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176490458 |
| 176490457 | 294.4000 | 294.173214 | C19H22N2O | [(1S,13R,14R)-15-ethylidene-3,17-diazapentacyclo[12.3.1.02,10.04,9.012,17]octadeca-2(10),4,6,8-tetraen-13-yl]methanol | 604-99-9 | InChI=1S/C19H22N2O/c1-2-11-9-21-17-8-14-12-5-3-4-6-16(12)20-19(14)18(21)7-13(11)15(17)10-22/h2-6,13,15,17-18,20,22H,7-10H2,1H3/t13-,15+,17?,18-/m0/s1 | VXTDUGOBAOLMED-KFMYLYIWSA-N | CC=C1CN2C3CC1C(C2CC4=C3NC5=CC=CC=C45)CO | True | False | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176490457 |
| 176490456 | 292.4000 | 274.016713 | C19H20N2O | (1S,13R,14R)-15-ethylidene-3,17-diazapentacyclo[12.3.1.02,10.04,9.012,17]octadeca-2(10),4,6,8-tetraene-13-carbaldehyde | C11634 | InChI=1S/C19H20N2O/c1-2-11-9-21-17-8-14-12-5-3-4-6-16(12)20-19(14)18(21)7-13(11)15(17)10-22/h2-6,10,13,15,17-18,20H,7-9H2,1H3/t13-,15+,17?,18-/m0/s1 | MHASSCPGKAMILD-KFMYLYIWSA-N | CC=C1CN2C3CC1C(C2CC4=C3NC5=CC=CC=C45)C=O | True | False | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176490456 |
| 176490455 | 292.4000 | 274.016713 | C19H20N2O | (1S,13S,14R)-15-ethylidene-3,17-diazapentacyclo[12.3.1.02,10.04,9.012,17]octadeca-2(10),4,6,8-tetraene-13-carbaldehyde | InChI=1S/C19H20N2O/c1-2-11-9-21-17-8-14-12-5-3-4-6-16(12)20-19(14)18(21)7-13(11)15(17)10-22/h2-6,10,13,15,17-18,20H,7-9H2,1H3/t13-,15-,17?,18-/m0/s1 | MHASSCPGKAMILD-FDGRXGLPSA-N | CC=C1CN2C3CC1C(C2CC4=C3NC5=CC=CC=C45)C=O | True | False | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=176490455 |
Database matching after adduct annotation with emzed annotate_adducts is straighforward since it is sufficient to match the database column m0 with the peak table column adduct_m0.
In case the peaks table was built with mzmine we can use the column isotope_base_peak_id to keep only the monoisotopic features (and the features without isotopologues):
[36]:
t = t_annotate.filter(
t_annotate.isotope_base_peak.is_none()
| (t_annotate.isotope_base_peak == t_annotate.id)
)
print(f"before: {len(t_annotate)}\tafter: {len(t)} ")
t.col_names
before: 311 after: 259
[36]:
('id',
'parent_id',
'mz',
'rt',
'mzmin',
'mzmax',
'rtmin',
'rtmax',
'area',
'height',
'width',
'peakmap',
'isotope_group_id',
'isotope_base_peak',
'isotope_group_size',
'isotope_charge',
'adduct_name',
'adduct_isotopes',
'adduct_isotopes_abundance',
'adduct_m0',
'adduct_cluster_id')
[37]:
# keep only features with KEGG match
t_match = t.join(kegg, t.adduct_m0.approx_equal(kegg.m0, 0.003, 0.0))
print(f"number of matches: {len(t_match)}")
t_match[:10]
number of matches: 548
[37]:
| id | parent_id | mz | rt | mzmin | mzmax | rtmin | rtmax | area | height | width | isotope_group_id | isotope_base_peak | isotope_group_size | isotope_charge | adduct_name | adduct_isotopes | adduct_isotopes_abundance | adduct_m0 | adduct_cluster_id | cid__0 | mw__0 | m0__0 | mf__0 | iupac__0 | synonyms__0 | inchi__0 | inchikey__0 | smiles__0 | is_in_kegg__0 | is_in_hmdb__0 | is_in_biocyc__0 | url__0 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | MzType | RtType | MzType | MzType | RtType | RtType | float | float | RtType | int | int | int | int | str | str | float | MzType | int | str | float | float | str | str | str | str | str | str | bool | bool | bool | str |
| 2382 | 2 | 86.059761 | 1.25 m | 86.059662 | 86.059853 | 1.22 m | 1.35 m | 6.63e+06 | 2.24e+06 | 0.13 m | 73 | 2382 | 3 | 1 | M+H | +[1]H | 0.999885 | 85.052485 | 0 | 6406 | 85.1000 | 85.052764 | C4H7NO | 2-hydroxy-2-methylpropanenitrile | ACETONE CYANOHYDRIN, 2-Hydroxy-2-methylpropanenitrile, 75-86-5, Acetone cyanhydrin, 2-Hydroxyisobutyronitrile, 2-Methyllactonitrile, 2-Cyano-2-propanol, alpha-Hydroxyisobutyronitrile, Acetoncyanhydrin, 2-Cyano-2-hydroxypropane, Acetoncianidrina, Acetonkyanhydrin, 2-Propanone, cyanohydrin, Lactonitrile, 2-methyl-, Propanenitrile, 2-hydroxy-2-methyl-, Acetoncianhidrinei, Acetoncyaanhydrine, Acetonecyanhydrine, Cyanhydrine d'acetone, USAF RH-8, 2-Hydroxy-2-methylpropionitrile, RCRA waste number P069, CO1YOV1KFI, DTXSID7025427, 2-hydroxy-2-methyl-propionitrile, acetone-cyanohydrin, 2-Cyanopropan-2-ol, CHEBI:15348, NSC-131093, DTXCID705427, acetoncyanhydrine, Cyanohydrin-2-propanone, RefChem:552382, 200-909-4, .alpha.-Hydroxyisobutyronitrile, NSC 131093, 2-hydroxy-2-methyl-propanenitrile, MFCD00004455, WLN: QX1&1&CN, Acetonkyanhydrin [Czech], Acetoncyanhydrin [German], Acetoncianidrina [Italian], Acetoncyaanhydrine [Dutch], Acetonecyanhydrine [French], CAS-75-86-5, Acetoncianhidrinei [Romanian], Acetone cyanohydrin, stabilized, CCRIS 4657, Cyanhydrine d'acetone [French], HSDB 971, a-hydroxyisobutyronitrile, UNII-CO1YOV1KFI, EINECS 200-909-4, UN1541, RCRA waste no. P069, BRN 0605391, AI3-04257, aceton cyanohydrin, acetone cyanhydrine, acetone cyanohydrine, acetone-cyanohydrine, acetone cyano-hydrin, 2-hydroxy-2-cyanopropane, Acetone cyanohydrin, 99%, EC 200-909-4, SCHEMBL34145, 4-03-00-00785 (Beilstein Handbook Reference), NSC977, SCHEMBL3827613, 2-methyl-2-hydroxypropionitrile, ACETONE CYANOHYDRIN [MI], CHEMBL1231861, DIMETHYLKETONE CYANOHYDRIN, 2-Methyl-2-hydroxypropanenitrile, NSC-977, NSC7080, 2-hydroxy-2-methylpropane nitrile, 2-methyl-2-oxidanyl-propanenitrile, 2-Hydroxy-2-methylpropanenitrile #, NSC-7080, Tox21_201490, Tox21_303256, BBL011349, NSC131093, SBB004397, STL146439, AKOS000118890, DB02203, UN 1541, NCGC00249054-01, NCGC00257016-01, NCGC00259041-01, M0361, NS00001430, EN300-19189, C02659, A838531, Q222936, Acetone cyanohydrin, stabilized [UN1541] [Poison], InChI=1/C4H7NO/c1-4(2,6)3-5/h6H,1-2H, F1908-0094 | InChI=1S/C4H7NO/c1-4(2,6)3-5/h6H,1-2H3 | MWFMGBPGAXYFAR-UHFFFAOYSA-N | CC(C)(C#N)O | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=6406 |
| 2391 | 15 | 94.044815 | 6.11 m | 94.044685 | 94.044945 | 6.09 m | 6.18 m | 3.34e+05 | 1.16e+05 | 0.09 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 76.010990 | 146 | 2723790 | 76.1200 | 76.009519 | CH4N2S | thiourea | THIOUREA, 62-56-6, Thiocarbamide, Isothiourea, 2-Thiourea, Pseudothiourea, Sulfourea, Thiuronium, Sulourea, 2-Thiopseudourea, Thiocarbonic acid diamide, Urea, thio-, beta-Thiopseudourea, Thiomocovina, Urea, 2-thio-, Tsizp 34, Pseudourea, 2-thio-, Thiocarbamid, Thioharnstoff, Thiokarbamid, USAF EK-497, carbonothioic diamide, RCRA waste number U219, CCRIS 588, DTXSID9021348, GYV9AM2QAG, H2NC(S)NH2, EPA Pesticide Chemical Code 080201, CHEBI:36946, AI3-03582, NSC-5033, DTXCID101348, NSC5033, thiourea; thiocarbamide, RefChem:6307, 200-543-5, Carbamimidothioic acid, Sulfouren, MFCD00008067, thio-urea, NSC 5033, 17356-08-0, .beta.-Thiopseudourea, (NH2)2CS, CHEMBL260876, TOU, Thiomocovina [Czech], Caswell No. 855, sulfocarbamide, RCRA waste no. U219, CAS-62-56-6, S C (N H2)2, HSDB 1401, THIOUREA, ACS, UNII-GYV9AM2QAG, EINECS 200-543-5, aminothioamide, thiopseudourea, aminothiocarboxamide, 2-Thio-Pseudourea, Thiocarbonic diamide, 2-Thio-Urea, beta -thiopseudourea, Thiourea, 99%, Sulfourea;Thiocarbamide, THIOUREA [HSDB], THIOUREA [IARC], WLN: ZYZUS, THIOUREA [MI], THIOUREA [VANDF], Urea, thio- (8CI), THIOUREA [WHO-DD], EC 200-543-5, SCHEMBL1008, Thiourea ACS Reagent Grade, SCHEMBL50650, Thiourea, LR, >=98%, MLS002454451, BIDD:ER0582, SCHEMBL192382, Propylthiouracil EP Impurity A, SCHEMBL4877346, SCHEMBL4877351, SCHEMBL9328938, SCHEMBL9794709, HMS2234E12, HMS3369M21, BCP27948, STR00054, Tox21_201873, Tox21_302767, BDBM50229993, STL194300, Thiourea, ACS reagent, >=99.0%, AKOS000269032, AKOS028109302, AT44959, CCG-207963, FT46947, UN 2877, Thiourea, ReagentPlus(R), >=99.0%, NCGC00091199-01, NCGC00091199-02, NCGC00091199-03, NCGC00256530-01, NCGC00259422-01, Thiourea, >=99.999% (metals basis), BP-31025, SMR000857187, Thiourea, JIS special grade, >=98.0%, Thiourea, p.a., ACS reagent, 99.0%, NS00002781, T0445, T2475, T2835, EN300-19634, PROPYLTHIOURACIL IMPURITY A [EP IMPURITY], 10.14272/UMGDCJDMYOKAJW-UHFFFAOYSA-N.1, A833853, F044741, Q528995, Thiourea, puriss. p.a., ACS reagent, >=99.0%, doi:10.14272/UMGDCJDMYOKAJW-UHFFFAOYSA-N.1, F0001-1650, Thiourea, Pharmaceutical Secondary Standard; Certified Reference Material | InChI=1S/CH4N2S/c2-1(3)4/h(H4,2,3,4) | UMGDCJDMYOKAJW-UHFFFAOYSA-N | C(=S)(N)N | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=2723790 |
| 2407 | 39 | 112.050262 | 1.63 m | 112.050102 | 112.050461 | 1.59 m | 1.72 m | 8.53e+05 | 2.85e+05 | 0.13 m | - | - | - | - | M+H | +[1]H | 0.999885 | 111.042986 | 2 | 68791 | 111.1000 | 111.043262 | C4H5N3O | 4-amino-1,3-diazabicyclo[3.1.0]hex-3-en-2-one | Imexon, 59643-91-3, DTXSID1046895, BM-06002, 4-Amino-1,3-diazabicyclo[3.1.0]hex-3-en-2-one, 4-Imino-1,3-diazabicyclo(3.1.0)hexan-2-one, NSC-313425, DTXCID9026895, 4-Imino-1,3-diazabicyclo[3.1.0]hexan-2-one, 4-Amino-1,3-diazabicyclo(3.1.0)hex-3-en-2-one, NSC714597, RefChem:790614, 4-imino-1,3-diazabicyclo(3,1,0)-hexan-2-one, 261-838-2, 8F63U28T2V, Amplimexon, Imexonum, MLS002702958, NSC313425, NCGC00181304-01, Imexon (USAN/INN), 4-Imino-1,3-diazabicyclo-[3.1.0]hexan-2-one, SCHEMBL154584, CHEMBL146428, GTPL8273, orb1707185, SCHEMBL3148692, Amplimexon (proposed trade name), HMS3264B05, Tox21_112779, AKOS006273819, AOP-990001, CCG-213631, DB05003, NSC-714597, NCGC00389454-01, NCI60_002705, SMR001566772, BM 06 002, CAS-59643-91-3, NS00034209, D08932, AB01013867_03, 4-Imino-1,3-diazabicyclo-[3.1.0]-hexan-2-one, 643I913, BRD-A78195072-001-06-2 | InChI=1S/C4H5N3O/c5-3-2-1-7(2)4(8)6-3/h2H,1H2,(H2,5,6,8) | BIXBBIPTYBJTRY-UHFFFAOYSA-N | C1C2N1C(=O)N=C2N | True | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=68791 |
| 2407 | 39 | 112.050262 | 1.63 m | 112.050102 | 112.050461 | 1.59 m | 1.72 m | 8.53e+05 | 2.85e+05 | 0.13 m | - | - | - | - | M+H | +[1]H | 0.999885 | 111.042986 | 2 | 597 | 111.1000 | 111.043262 | C4H5N3O | 6-amino-1H-pyrimidin-2-one | cytosine, 71-30-7, 4-Amino-2-hydroxypyrimidine, 4-aminopyrimidin-2(1H)-one, Cytosinimine, 2(1H)-Pyrimidinone, 4-amino-, 4-Amino-2(1H)-pyrimidinone, 6-amino-1H-pyrimidin-2-one, DTXSID4044456, CHEBI:16040, NSC-27787, 8J337D1HZY, 4-amino-3h-pyrimidin-2-one, DTXCID2024456, NSC27787, 6-amino-1,2-dihydropyrimidin-2-one, RefChem:5728, 200-749-5, 4-Aminouracil, 6-Aminopyrimidin-2(1h)-One, MFCD00006034, 4-aminopyrimidin-2-ol, Cytosin, 2(1H)-pyrimidinone, 6-amino-, Cyt, 4-amino-2-oxo-1,2-dihydropyrimidine, 4-amino-2-pyrimidinol, Zytosin, AI3-52281, 107646-83-3, 134434-40-5, 107646-84-4, 134434-39-2, 2(1H)-Pyrimidinone, 3,4-dihydro-4-imino-, (E)- (9CI), 4-Aminopyrimidin-2-(1H)-one, 2(1H)-Pyrimidinone, 3,4-dihydro-4-imino-, (Z)- (9CI), 4-Aminopyrimidin-2(1H)-one (Cytosine), Cytosine-5,6-d2, 6-amino-3-hydropyrimidin-2-one, pyrimidine, 4-amino-2-hydroxy-, SMR000857094, 4-Amino-1H-pyrimidin-2-one, EINECS 200-749-5, Lamivudine EP Impurity E, NSC 27787, 2-Pyrimidinol, 1,4-dihydro-4-imino-, aminopyrimidone, UNII-8J337D1HZY, iminopyrimidinone, 3h-cytosine, 66460-18-2, 66460-21-7, Cytosine (8CI), 4-amino-1,2-dihydropyrimidin-2-one, Cytosine (Standard), Lamivudine impurity c, 2-Pyrimidinol, 1,4-dihydro-4-imino-, (Z)- (9CI), Gemcitabine impurity A, Cytosine, >=99%, 2(1H)-Pyrimidinone, 4-amino- (9CI), Lamivudine impurity c rs, CYTOSINE [MI], 4-amino-pyrimidin-2-ol, 4-Amino-2-oxypyrimidine, 2(1H)-Pyrimidinone, 3,4-dihydro-4-imino-, bmse000180, CYTOSINE [USP-RS], CYTOSINE [WHO-DD], Epitope ID:167475, 4-Amino-2(1H)pyrimidone, EC 200-749-5, SCHEMBL4059, SCHEMBL8618, 4-Amino-2(1H)-pyrimidone, 2-Hydroxy-6-amino-pyrimidin, MLS001332635, MLS001332636, CHEMBL15913, SCHEMBL308339, SCHEMBL437958, SCHEMBL437959, SCHEMBL655341, 2-Pyrimidinol, 1,6-dihydro-6-imino-, (E)- (9CI), GTPL8490, orb1302527, SCHEMBL4818561, SCHEMBL6547006, SCHEMBL8921275, SCHEMBL10402537, HY-I0626R, MSK6920, HMS2233N21, HMS3369N05, HMS5083D22, Cytosine, >=99.0% (HPLC), ALBB-021996, BCP22793, HY-I0626, STR01426, Tox21_302139, s4893, SBB071711, STK366767, STL455080, AKOS000120336, AKOS005443393, AKOS015896942, AC-2489, BCP9000005, CCG-266052, CS-W020703, FC03781, 2-Pyrimidinol, 1,6-dihydro-6-imino-, CAS-71-30-7, CID 5274263, SRI-2354-05, NCGC00247019-01, NCGC00255926-01, 66398-98-9, 66460-19-3, BP-20183, Cytosine, Vetec(TM) reagent grade, 99%, NCI60_012445, SY001643, 4-imino-3,4-dihydropyrimidin-2(1H)-one, DB-029615, DB-268893, DB-268963, DB-271170, DB-271171, DB-295125, LAMIVUDINE IMPURITY E [EP IMPURITY], LAMIVUDINE IMPURITY C [USP IMPURITY], NS00006844, ST45026865, EN300-21504, 71C307, C00380, 006C034, 2-Pyrimidinol,1,6-dihydro-6-imino-,(E)-(9ci), F036359, Q178425, 2(1H)-Pyrimidinone,3,4-dihydro-4-imino-,(E)-(9ci), CBA1D098-C5AB-46CE-AAC6-754572886EB2, Z203045338, Cytosine, United States Pharmacopeia (USP) Reference Standard, Cytosine, Pharmaceutical Secondary Standard; Certified Reference Material, Gemcitabine impurity A, European Pharmacopoeia (EP) Reference Standard, 66460-16-0 | InChI=1S/C4H5N3O/c5-3-1-2-6-4(8)7-3/h1-2H,(H3,5,6,7,8) | OPTASPLRGRRNAP-UHFFFAOYSA-N | C1=C(NC(=O)N=C1)N | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=597 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 125468 | 100.1200 | 100.052430 | C5H8O2 | (E)-2-methylbut-2-enoic acid | TIGLIC ACID, 80-59-1, (E)-2-Methylbut-2-enoic acid, Tiglinic acid, trans-2,3-Dimethylacrylic acid, trans-2-Methyl-2-butenoic acid, trans-2-Methylcrotonic acid, (E)-2-Methyl-2-butenoic acid, (E)-2,3-Dimethylacrylic acid, (E)-2-Methylcrotonic acid, 2-Butenoic acid, 2-methyl-, (2E)-, Crotonic acid, 2-methyl-, (E)-, 2-Butenoic acid, 2-methyl-, (E)-, 2,3-Dimethylacrylic acid, (E)-, FEMA No. 3599, trans-alpha,beta-Dimethylacrylic acid, 2-methyl-2E-butenoic acid, CHEBI:9592, I5792N03HC, NSC-8999, NSC-44235, DTXSID80883257, RefChem:25822, (e)-alpha-methylcrotonic acid, DTXCID201022801, 201-295-0, Cevadic acid, 2-Methyl-2-butenoic acid, 2-methylbut-2-enoic acid, tiglate, 13201-46-2, alpha-Methylcrotonic acid, MFCD00066864, (2E)-2-methylbut-2-enoic acid, 2-Methylcrotonic acid, NSC 44235, (E)-2-methyl-Crotonic acid, 2,3-Dimethylacrylic acid, trans-.alpha.,.beta.-Dimethylacrylic acid, 2-methyl-(E)-2-butenoic acid, alpha,beta-dimethyl acrylic acid, NSC8999, NSC44235, Tiglicacid, methyl methacrylic acid, alpha-Methylcrotonic acid, (E)-, 2-Methyl-2-butenoic acid, (E)-, EINECS 201-295-0, 2-Butenoic acid,2-methyl-, BRN 1236500, Tiglinsaeure, Cevadate, Tiglinate, epsilon-Tiglate, UNII-I5792N03HC, AI3-36118, E-Tiglate, Methylbutenoicacid, E-Tiglic acid, HSDB 7614, (2E)-2-Methyl-2-butenoic acid, 2-methyl-Crotonate, epsilon-Tiglic acid, EINECS 236-167-3, methyl crotonic acid, methylmethacrylic acid, 2,3-Dimethylacrylate, 2-methyl-2-butenoate, Tiglic acid, (E)-, 2-Methylbut-2-enoate, 2-methyl-Crotonic acid, trans-2-Methylcrotonate, (E)-2-Methylcrotonate, 2-methylbut-2-enoicacid, (E)-2-methyl-Crotonate, alpha-methyl-crotonic acid, TIGLIC ACID [MI], trans-2,3-Dimethylacrylic acid (Tiglic acid), bmse000727, TIGLIC ACID [HSDB], trans-2,3-Dimethylacrylate, (E)-2,3-Dimethylacrylate, 2-methyl-(E)-2-butenoate, trans-2-Methyl-2-butenoate, (E)-2-methyl-2-Butenoate, SCHEMBL18814, 4-02-00-01552 (Beilstein Handbook Reference), CHEMBL52416, SCHEMBL487578, (2E)-2-Methyl-2-butenoate, GTPL6499, orb1301709, SCHEMBL11874672, SCHEMBL27864425, trans-Crotonic acid, 2-methyl-, CHEBI:36432, trans-alpha,beta-Dimethylacrylate, E)-2-METHYLCROTONIC ACID, trans-2-methyl-but-2-enoic acid, BCP18945, (2E)-2-Methyl-2-butenoic acid #, LMFA01020030, s3789, AKOS003375681, CCG-266028, CS-W013715, FM71312, HY-W012999, trans-2,3-Dimethylacrylic acid, 98%, (E)-.ALPHA.-METHYLCROTONIC ACID, NCGC00648718-02, DA-58577, LS-13047, SY015876, 2-METHYLBUT-2-ENOIC ACID, (E)-, METHYL-2-BUTENOIC ACID, TRANS-2-, CS-0356606, NS00009823, T0246, Tiglic acid/trans-2,3-Dimethylacrylic acid), EN300-83150, C08279, D78012, EN300-370249, TRANS-2-METHYL-2-BUTENOIC ACID [FHFI], trans-2-Methyl-2-butenoic acid, >=99%, FG, F545226, F552761, Q425475, Q27116830, F0001-2086, Z1205493556, trans-2-Methylcrotonic acid = trans-2-Methyl-2-butenoate, trans-2-Methylcrotonic acid = trans-2-Methyl-2-butenoic acid, trans-2-Methylcrotonic acid = trans-2-Methyl-2-butenoic acid = Tiglinate, alpha,beta-dimethyl acrylic acid; 2-Methyl-2-butenoic acid; (E)-2-methyl-Crotonic acid, trans-2-Methylcrotonic acid = trans-2-Methyl-2-butenoic acid = Tiglinic acid, trans-2,3-Dimethylacrylic acid; trans-2-Methyl-2-butenoic acid; (2E)-2-Methyl-2-butenoic acid | InChI=1S/C5H8O2/c1-3-4(2)5(6)7/h3H,1-2H3,(H,6,7)/b4-3+ | UIERETOOQGIECD-ONEGZZNKSA-N | CC=C(C)C(=O)O | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=125468 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 31261 | 100.1200 | 100.052430 | C5H8O2 | pentane-2,4-dione | Acetylacetone, 2,4-Pentanedione, Pentane-2,4-dione, 123-54-6, Acetoacetone, ACAC, 2,4-Dioxopentane, Diacetylmethane, ACETYL ACETONE, Pentanedione, Pentan-2,4-dione, Pentanedione-2,4, Acetyl 2-propanone, Acetone, acetyl-, Hacac, 2-Propanone, acetyl-, DTXSID4021979, CHEBI:14750, 46R950BP4J, NSC-5575, DTXCID601979, RefChem:5511, GlyTouCan:G58526LS, G58526LS, 204-634-0, 2,4-Pentadione, 2,4-Pentandione, acetylaceton, acetyl-acetone, NSC 5575, MFCD00008787, CH3COCH2COCH3, 4-Hydroxy-3-penten-2-one, CH3-CO-CH2-CO-CH3, CCRIS 3466, HSDB 2064, 14024-62-5, EINECS 204-634-0, UN2310, BRN 0741937, UNII-46R950BP4J, AI3-02266, 81235-32-7, pentane-2, pentan-2, acetylacetone (2,4-pentanedione), 2,4 pentanedione, 2.4-pentanedione, pentane2,4-dione, Acetyl-2-Propanone, 2,4-pentane-dione, ACETYLACETONE ENOL, ACETYLACETONE [MI], 1-methylbutane-1,3-dione, EC 204-634-0, SCHEMBL1608, NCIOpen2_000702, 4-01-00-03662 (Beilstein Handbook Reference), Pentane-2,4-dione [UN2310] [Flammable liquid], ACETYL ACETONE [HSDB], SCHEMBL270911, SCHEMBL379482, CHEMBL191625, SCHEMBL9256650, WLN: 1V1V1, SCHEMBL10698605, Acetylacetone;Pentane-2,4-dione, BDBM22766, NSC5575, Acetylacetone, analytical standard, BCP31333, STR00020, Tox21_200414, LMFA12000075, MSK161365, SBB009914, AKOS000118994, FP35056, UN 2310, Acetylacetone, ReagentPlus(R), >=99%, NCGC00248599-01, NCGC00257968-01, BP-30252, CAS-123-54-6, PD193123, ST059915, Acetylacetone, JIS special grade, >=99%, DB-020012, DB-318551, DS-002710, NS00007112, P0052, EN300-19143, F100600, Q413447, Pentane-2,4-dione [UN2310] [Flammable liquid], F1908-0168, InChI=1/C5H8O2/c1-4(6)3-5(2)7/h3H2,1-2H, Acetylacetone, produced by Wacker Chemie AG, Burghausen, Germany, >=99.5% (GC) | InChI=1S/C5H8O2/c1-4(6)3-5(2)7/h3H2,1-2H3 | YRKCREAYFQTBPV-UHFFFAOYSA-N | CC(=O)CC(=O)C | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=31261 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 10953 | 100.1200 | 100.052430 | C5H8O2 | oxan-2-one | delta-Valerolactone, 542-28-9, TETRAHYDRO-2H-PYRAN-2-ONE, 5-Valerolactone, 2H-Pyran-2-one, tetrahydro-, DTXSID6044438, CHEBI:16545, NSC-6247, 14V1X9149L, DTXCID4024438, RefChem:585696, 208-807-1, oxan-2-one, tetrahydropyran-2-one, Tetrahydro-2-pyranone, .delta.-Valerolactone, Valerolactone, 26354-94-9, .delta.-Valeryllactone, tetrahydro-pyran-2-one, 1-Oxacyclohexan-2-one, NSC 6247, MFCD00006645, CHEMBL452383, Penta-1,5-lactone, 3H-4,5,6-trihydropyran-2-one, delta-Valeryllactone, d-valerolactone, Cyclopentanolide, 5-pentanolide, UNII-14V1X9149L, Pentan-5-olide, Delta valerolactone, Delta-Valerolactotie, EINECS 208-807-1, delta -Valerolactone, .delta.-Pentalactone, tetrahydro-4H-pyranone, AI3-25024, Pentanoic acid, 5-hydroxy-, delta-lactone, Valeric acid, delta-hydroxy-, delta-lactone, 98% may contain polymer, WLN: T6OVTJ, EC 208-807-1, SCHEMBL37722, SCHEMBL165797, SCHEMBL893251, Valeric acid, .delta.-lactone, orb1302537, SCHEMBL1417857, SCHEMBL2175853, SCHEMBL3111979, SCHEMBL16968705, SCHEMBL28683143, Pentanoic acid, .delta.-lactone, NSC6247, NSC65442, STR08736, delta-Valerolactone, technical grade, Tox21_302166, BDBM50360797, NSC 65442, NSC-65442, s3099, SBB060349, AKOS009158729, CS-W013713, FV62738, HY-W012997, MSK169937-100B, 5-Hydroxypentanoic acid .delta.-lactone, MSK169937-1000B, NCGC00255756-01, BP-14040, CAS-542-28-9, Delta-valerolactone (may contain polymer), SY018264, DB-050659, NS00005148, ST51046546, V0039, EN300-43042, Pentanoic acid, 5-hydroxy-, .delta.-lactone, tetrahydro-2H-pyran-2-one,2-Perhydropyranone, C02240, delta-Valerolactone Solution in Acetone, 100ug/mL, F448260, Q903610, Valeric acid, .delta.-hydroxy-, .delta.-lactone, delta-Valerolactone Solution in Acetone, 1000ug/mL, InChI=1/C5H8O2/c6-5-3-1-2-4-7-5/h1-4H, Z432085120 | InChI=1S/C5H8O2/c6-5-3-1-2-4-7-5/h1-4H2 | OZJPLYNZGCXSJM-UHFFFAOYSA-N | C1CCOC(=O)C1 | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=10953 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 8821 | 100.1200 | 100.052430 | C5H8O2 | ethyl prop-2-enoate | ETHYL ACRYLATE, 140-88-5, Acrylic acid ethyl ester, 2-Propenoic acid, ethyl ester, Ethyl propenoate, Ethyl 2-propenoate, Ethylacrylaat, Ethylakrylat, Etil acrilato, Ethyl acrylate, inhibited, Acrylic acid, ethyl ester, Aethylacrylat, Etilacrilatului, Akrylanem etylu, Carboset 511, Ethoxycarbonylethylene, Acrylate d'ethyle, Acrylsaeureaethylester, Ethyl acrylate (inhibited), RCRA waste number U113, Ethyl propenoate, inhibited, Acrylic acid, ethyl ester (inhibited), Ethylester kyseliny akrylove, NCI-C50384, 2-Propenoic Acid Ethyl Ester, FEMA No. 2418, FEMA Number 2418, DTXSID4020583, NSC-8263, 71E6178C9T, DTXCID00583, Ethyl acrylic ester, RefChem:7348, 205-438-8, 64050-18-6, ethyl prop-2-enoate, Etilacrilatului [Romanian], NSC 8263, ethyl 2E-propenoate, Acrylic acid-ethyl ester, Etilacrilatului(roumanian), MFCD00009188, CH2=CHCOOC2H5, Ethyl Acrylate (stabilized with MEHQ), ETHYL-D5 ACRYLATE, CHEBI:82327, Ethyl ester of 2-propenoic acid, WE(2:0/3:1(2E)), Ethyl Acrylate (Stabilized with 4-methoxyphenol), Ethylakrylat [Czech], Ethylacrylaat [Dutch], Aethylacrylat [German], Ethylacrylate, Etil acrilato [Italian], Ethyl acrylate (natural), Akrylanem etylu [Polish], Acrylate d'ethyle [French], CCRIS 248, Acrylsaeureaethylester [German], HSDB 193, EINECS 205-438-8, Ethylester kyseliny akrylove [Czech], UN1917, RCRA waste no. U113, BRN 0773866, AI3-15734, UNII-71E6178C9T, EC 205-438-8, SCHEMBL3180, ETHYL ACRYLATE [MI], SCHEMBL35914, ETHYL ACRYLATE [FCC], 4-02-00-01460 (Beilstein Handbook Reference), CHEMBL52084, ETHYL ACRYLATE [FHFI], ETHYL ACRYLATE [HSDB], ETHYL ACRYLATE [IARC], SCHEMBL293486, SCHEMBL294547, SCHEMBL525535, WLN: 2OV1U1, Ethyl acrylate, inhibited [UN1917] [Flammable liquid], FEMA 2418, FEMA-2418, NSC8263, Ethyl acrylate, analytical standard, BCP06341, Tox21_202513, BBL011497, LMFA07010505, SBB060348, STL146609, Ethyl Acrylate, stabilized with MEHQ, AKOS005721113, NCGC00091041-01, NCGC00260062-01, CAS-140-88-5, VS-02962, Propenoic acid,ethyl ester (ethylacrylate), DB-042537, Ethyl Acrylate 1000 microg/mL in Methanol, A0143, Ethyl acrylate, >=99.5%, stabilized, FG, NS00006376, ST51046544, EN300-19964, Ethyl acrylate, SAJ first grade, >=99.0%, C19238, Ethyl Acrylate stabilized with 10 - 20 ppm MEHQ, F094594, Q343014, PROPENOIC ACID,ETHYL ESTER (ETHYLACRYLATE), Ethyl acrylate, inhibited [UN1917] [Flammable liquid], F1908-0175, Ethyl acrylate, contains 10-20 ppm MEHQ as inhibitor, 99%, InChI=1/C5H8O2/c1-3-5(6)7-4-2/h3H,1,4H2,2H, 2-Propenoic acid, 1,1'-((dihydro-5-(2-hydroxyethyl)-2,4,6-trioxo-1,3,5-triazine-1,3(2H,4H)-diyl)di-2,1-ethanediyl) ester | InChI=1S/C5H8O2/c1-3-5(6)7-4-2/h3H,1,4H2,2H3 | JIGUQPWFLRLWPJ-UHFFFAOYSA-N | CCOC(=O)C=C | True | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=8821 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 6658 | 100.1200 | 100.052430 | C5H8O2 | methyl 2-methylprop-2-enoate | METHYL METHACRYLATE, 80-62-6, methyl 2-methylprop-2-enoate, Methylmethacrylate, Methyl methylacrylate, Methyl 2-methylpropenoate, Methacrylic acid methyl ester, Pegalan, Methyl-methacrylat, Diakon, Methyl 2-methyl-2-propenoate, Acryester M, Methacrylate de methyle, Methyl 2-methylacrylate, 2-Propenoic acid, 2-methyl-, methyl ester, Metakrylan metylu, Metil metacrilato, Methacrylsaeuremethyl ester, Methylmethacrylaat, 2-(Methoxycarbonyl)-1-propene, 2-Methyl-2-propenoic acid methyl ester, Rcra waste number U162, Methyl alpha-methylacrylate, Methyl methacrylate monomer, NCI-C50680, Methacrylic acid, methyl ester, Acrylic acid, 2-methyl-, methyl ester, Monocite methacrylate monomer, Methylester kyseliny methakrylove, CHEBI:34840, 2-methylacrylic acid methyl ester, DTXSID2020844, NSC-4769, DTXCID80844, 196OC77688, NSC4769, Cranioplasts, Kallocryls, Metaplices, Sintices, Methacrylate Methyl Monomer, Methacrylate monomer, Zimmer Bone Cements, Bone Cement, Zimmer, Cement, Zimmer Bone, Bone Cements, Zimmer, Cements, Zimmer Bone, Methylmethacrylate Methyl Monomer, Methacrylate Methyl Monomers, Methyl Monomer, Methacrylate, Monomer, Methacrylate Methyl, Methyl Monomers, Methacrylate, methyl-2-methyl-2-propenoate, Monomers, Methacrylate Methyl, Methyl ester of methacrylic acid, Methylmethacrylate Methyl Monomers, Methyl Monomer, Methylmethacrylate, Monomer, Methylmethacrylate Methyl, Methyl Monomers, Methylmethacrylate, Monomers, Methylmethacrylate Methyl, 2-methylmethacrylate, RefChem:817845, 2-(Methoxycarbonyl)propene, 2-Methyl Acrylic Acid Methyl Ester, 2-Propenoic acid, 2-methyl-methyl ester, 201-297-1, TEB 3K, 2-Methylacrylic acid, methyl ester, 2-Methyl-acrylic acid methyl ester, NSC 4769, MFCD00008587, Methacrylic acid-methyl ester, Methyl .alpha.-methylacrylate, Cranioplast, Metaplex, Kallocryl A, Simplex P, Methyl methacrylate monomer, inhibited, 143476-91-9, Methyl Methacrylate (stabilized with 6-tert-Butyl-2,4-xylenol), Methyl ester of 2-methyl-2-propenoic acid, 114512-63-9, 51391-19-6, Methylmethacrylaat [Dutch], Metakrylan metylu [Polish], Methyl-methacrylat [German], Metil metacrilato [Italian], 9065-11-6, CCRIS 1364, HSDB 195, Methacrylate de methyle [French], Methacrylsaeuremethyl ester [German], EINECS 201-297-1, UN1247, RCRA waste no. U162, BRN 0605459, Plexiglass, Eudragit, Methylester kyseliny methakrylove [Czech], AI3-24946, methoxymethacrolein, UNII-196OC77688, MMA (stabilized), J69, Acrylic resins (PMMA), METHYL METHACTRYLATE, Epitope ID:131321, Methyl 2-methylacrylate #, Methyl methacrylate (MMA), EC 201-297-1, Methyl-.alpha.-methacrylate, SCHEMBL1849, CH2=C(CH3)COOCH3, 4-02-00-01519 (Beilstein Handbook Reference), NA 1247 (Salt/Mix), UN 1247 (Salt/Mix), BIDD:ER0634, CHEMBL49996, Methyl methacrylate, 99.5%, SCHEMBL217190, SCHEMBL295831, SCHEMBL865215, WLN: 1UY1&VO1, Methyl methacrylate, stabilized, 'monocite' Methacrylate monomer, SCHEMBL3893737, SCHEMBL9375987, SCHEMBL10636466, Methyl methacrylate, CP,98.0%, METHYL METHACRYLATE [HSDB], METHYL METHACRYLATE [IARC], METHYLMETHACRYLATE [MART.], METHYL METHACRYLATE [VANDF], METHYLMETHACRYLATE [WHO-DD], Tox21_200367, MSK002111, SBB060556, STL283952, AKOS000120216, Methyl methacrylate, 99%, stabilized, FM34647, CAS-80-62-6, NCGC00091089-01, NCGC00091089-02, NCGC00257921-01, Methacrylic Acid Methyl Ester (stabilized, METHACRYLIC ACID METHYL ESTER [MI], DB-013559, CS-0503657, M0087, METHYL 2-METHYL-2-PROPENOATE [FHFI], NS00009302, ST51046719, EN300-19210, C19504, Methyl methacrylate 1000 microg/mL in Methanol, Methyl methacrylate, SAJ first grade, >=99.0%, A839957, F995248, Q382897, F0001-2087, InChI=1/C5H8O2/c1-4(2)5(6)7-3/h1H2,2-3H, Methacrylic acid-methyl ester 100 microg/mL in Cyclohexane, Methyl Methacrylate, (stabilized with 6-tert-Butyl-2,4-xylenol), Methyl methacrylate, contains <=30 ppm MEHQ as inhibitor, 99%, Methyl methacrylate, European Pharmacopoeia (EP) Reference Standard, Methyl methacrylate (MMA), 99.5%(GC), contains 30ppm MEHQ as stabilizer, Methyl methacrylate (MMA), AR, 99.0%, contains 30ppm MEHQ as stabilizer, Methyl methacrylate monomer, inhibited [UN1247] [Flammable liquid], PROPENOIC ACID,2-METHYL,METHYLESTER (METHACRYLATE METHYLESTER), 97555-82-3 | InChI=1S/C5H8O2/c1-4(2)5(6)7-3/h1H2,2-3H3 | VVQNEPGJFQJSBK-UHFFFAOYSA-N | CC(=C)C(=O)OC | True | True | False | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=6658 |
| 2418 | 57 | 118.085960 | 1.65 m | 118.085831 | 118.086128 | 1.62 m | 1.72 m | 7.23e+05 | 2.20e+05 | 0.10 m | - | - | - | - | M+NH4 | +[1]H4 [14]N | 0.995862 | 100.052135 | 4 | 3485 | 100.1200 | 100.052430 | C5H8O2 | pentanedial | glutaraldehyde, Pentanedial, 111-30-8, Glutaral, Glutaric dialdehyde, 1,5-Pentanedial, Sonacide, Glutardialdehyde, Glutaric acid dialdehyde, Glutaric aldehyde, Glutaraldehyd, Glutaralum, Aldesan, Alhydex, Ucarcide, Hospex, 1,3-Diformylpropane, 1,5-Pentanedione, Aldesen, Sterihyde L, Aldehyd glutarowy, Glutaclean, Sterihyde, NCI-C55425, Relugan GT, Relugan GTW, Glutarex 28, Cidex 7, Ucarcide 250, Relugan GT 50, Dioxopentane, Verucasep, DTXSID6025355, Coldcide-25 microbiocide, NSC-13392, Potentiated acid glutaraldehyde, CHEBI:64276, T3C89M417N, DTXCID605355, Panavirocide, Glutohyde, Pond Health Guard, RefChem:57623, GlyTouCan:G47927JT, G47927JT, 203-856-5, Cidex, Glutarol, Pentane-1,5-dial, Gluteraldehyde, Novaruca, Sporicidin, Aqucar, Veruca-sep, component of Cidex, MFCD00007025, NSC 13392, Sonacide (TN), Sterihyde L (TN), Glutaral (JAN/USP/INN), Glutaraldehyde (24-26% in Water), NCGC00091110-01, PTD, Glutaraldehyde (50% in water), Caswell No. 468, Glutaraldehyd [Czech], 1,3-Diformyl propane, Diswart, Gludesin, Glutaralum [INN-Latin], Glutarol-1,5-pentanedial, Aldehyd glutarowy [Polish], Glutaral [USAN:INN:JAN], CAS-111-30-8, CCRIS 3800, HSDB 949, EINECS 203-856-5, EPA Pesticide Chemical Code 043901, BRN 0605390, pentandial, Glutural, Ucarset, Virsal, UNII-T3C89M417N, Glutaral(usan), glutaric dihydride, Glutaral [USAN:USP:INN:JAN], GLUTARALDEHYDE, 25% SOLN, Bactron K31, Ucarcide 225, MINOXIDIL_met005, GLUTARAL [HSDB], GLUTARAL [USAN], pentane-1,5-dialdehyde, GLUTARAL [INN], GLUTARAL [JAN], Glutaral, INN, USAN, GLUTARAL [MART.], Protectol GDA, GT 50, SCHEMBL836, WLN: VH3VH, DIPYRIDAMOLE_met034, GLUTARAL [WHO-DD], EC 203-856-5, GLUTARALDEHYDE [MI], Pentane-1,5-dial solution, GLUTARALDEHYDE [FCC], SCHEMBL97707, 4-01-00-03659 (Beilstein Handbook Reference), BIDD:ER0299, GLUTARALDEHYDE [VANDF], orb2939722, SCHEMBL3145412, SCHEMBL5611212, SCHEMBL6464083, GLUTARAL [USP IMPURITY], CHEMBL1235482, SCHEMBL10422822, SCHEMBL10720109, MSK2528, Bio1_000462, Bio1_000951, Bio1_001440, Glutaraldehyde solution, 25% w/w, Glutaraldehyde solution, 50% w/w, Glutaraldehyde solution, 70% w/w, NSC13392, STR01121, Tox21_111083, Tox21_201742, Tox21_303295, STL281872, Gluteraldehyde Solution, 1000 mg/L, AKOS008967285, Glutaraldehyde (50per cent in water), DB03266, FG32551, Glutaric dialdehyde, 25%sol. In water, Glutaric dialdehyde, 25% sol. in water, NCGC00091110-02, NCGC00091110-03, NCGC00257231-01, NCGC00259291-01, DA-53608, G0067, G0068, Glutaraldehyde - 50 wt. % aqueous solution, NS00004136, EN300-18037, D01120, A802339, Q416475, Glutaraldehyde (ca. 50% in Water, ca. 5.6mol/L), Z57127529, F2191-0161 | InChI=1S/C5H8O2/c6-4-2-1-3-5-7/h4-5H,1-3H2 | SXRSQZLOMIGNAQ-UHFFFAOYSA-N | C(CC=O)CC=O | True | True | True | http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=3485 |
Note the number of matches does not correspond to the number of identified peaks:
[38]:
identified = len(set(t_match.id))
total = len(t)
print(f"identified: {identified/total*100:.1f}%")
identified: 38.6%
© Copyright 2012-2025 ETH Zurich
Last build 2025-10-28 13:23:38.200852.
Created using Sphinx 8.1.3.