[1]:
%%capture
# Setup
import emzed
import os
# path to files
data_folder = os.path.join(os.getcwd(), "tutorial_data")
path_ms2 = os.path.join(data_folder, "t_attach_ms2.table")
path_pm_ms2 = os.path.join(data_folder, "ms_prm.mzML")
# files
t_ms2 = emzed.io.load_table(path_ms2)
peakmap_ms2 = emzed.io.load_peak_map(path_pm_ms2)
# emzed.run_feature_finder_metabo parameter setup (UPLC-LTQ-Orbitrap data)
ff_params = dict(
common_chrom_peak_snr=10.0,
common_chrom_fwhm=3.0,
mtd_noise_threshold_int=7000.0,
mtd_mass_error_ppm=20.0,
mtd_reestimate_mt_sd="true",
mtd_trace_termination_criterion="outlier",
mtd_trace_termination_outliers=4,
mtd_min_sample_rate=0.5,
mtd_min_trace_length=5.0,
mtd_max_trace_length=-1.0,
epdet_width_filtering="auto",
epdet_masstrace_snr_filtering="false",
ffm_local_rt_range=2.0,
ffm_local_mz_range=5.0,
ffm_charge_lower_bound=0,
ffm_charge_upper_bound=3,
ffm_report_summed_ints="false",
ffm_isotope_filtering_model="none",
ffm_use_smoothed_intensities="false",
)
LC-MS level 1 approaches are often combined with tandem mass spectrometry yielding one or several fragment spectra per selected LC-MS peak to allow compound confirmation or identification. Selection of parent-ions can be targeted by i.e. a peak inclusion lists or untargeted by i.e. Top N approaches selecting the N most intense m/z signals of ms level 1 spectrum for fragmentation. The emzed ms2 module features assigning fragmentation spectra to their corresponding LC-MS level one peaks with
function attach_ms2_spectra
emzed.ms2.attach_ms2_spectra(peak_table, mode='union', mz_tolerance=0.0013, print_final_report=True,)
with arguments
peak_table: emzed Table defining LC-MS level 1 peaks checked for presence of fragmentation spectra with required columns “rtmin”, “rtmax”, “mzmin”, “mzmax” and “peakmap”.
mode: defines the representation of attached spectra. Possible modes are:
all: extracts a list of all ms2 spectra in within peak window.
max_range, c)max_energie: extract a single spectrum with widest m/z range (max_range) or with highest spectral intensity (max_energy)
d)union, e)intersection: merge spectra within peak window. Union results a spectrum with all peaks whereas intersection contains only peaks present in all spectra. Default value: union.
mz_tolerance: maximal allowed mz difference of two spectral peaks to be merged. Parameter is only needed for modes `union” and “intersection”.
print_final_report: prints some final report with diagnosis messages for testing if mz_tolerance parameter fits, i.e. you should set this parameter to True if you are not sure if mz_tolerance fits to your machines mass resolution. Default value is True.
returns in_place
In the following exmaples we will do show 2 possible use cases for the same data set. The first example is based on an inclusion list defining the fragmentation peaks. We can build a peak_table based on the inclusion list, which is provided
[2]:
t_ms2
[2]:
| id | precursor_mz | mzmin | mzmax | rtmin | rtmax |
|---|---|---|---|---|---|
| int | MzType | MzType | MzType | RtType | RtType |
| 0 | 465.098000 | 465.095000 | 465.101000 | 1.68 m | 1.95 m |
| 1 | 346.090000 | 346.087000 | 346.093000 | 0.93 m | 1.01 m |
| 2 | 201.048000 | 201.045000 | 201.051000 | 0.48 m | 0.59 m |
| 3 | 524.138000 | 524.135000 | 524.141000 | 1.43 m | 1.54 m |
The table defines for LC-MS level 1 peaks. We can now attach corresponding ms2 spectra to those peaks using the default mode union :
[3]:
%%capture
emzed.ms2.attach_ms2_spectra(t_ms2, mz_tolerance=0.003)
[4]:
t_ms2
[4]:
| id | precursor_mz | mzmin | mzmax | rtmin | rtmax | spectra_ms2 | num_spectra_ms2 | ms2_extraction_info |
|---|---|---|---|---|---|---|---|---|
| int | MzType | MzType | MzType | RtType | RtType | object | int | str |
| 0 | 465.098000 | 465.095000 | 465.101000 | 1.68 m | 1.95 m | [ |
38 | union |
| 1 | 346.090000 | 346.087000 | 346.093000 | 0.93 m | 1.01 m | [ |
11 | union |
| 2 | 201.048000 | 201.045000 | 201.051000 | 0.48 m | 0.59 m | [ |
30 | union |
| 3 | 524.138000 | 524.135000 | 524.141000 | 1.43 m | 1.54 m | [ |
10 | union |
The resulting table has now 3 additional columns spectra_ms2 cotaining the merged spectrum, num_spectra_ms2 with the number of merged spectra, and ms2_extraction_info showing applied mode When we now open the table with the table explorer we can visualize the fragmentation spectra.
emzed.gui.inspect(t_ms2)
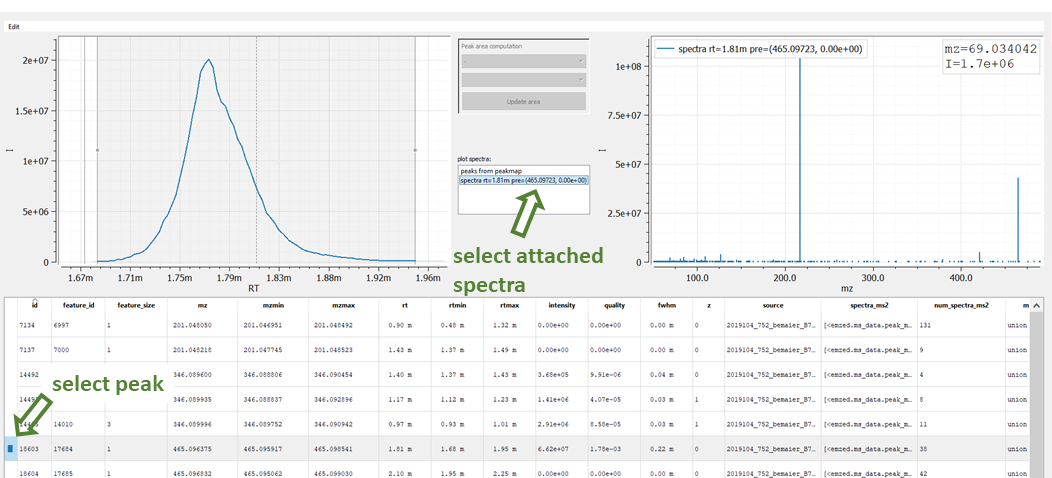
To show a fragmentation spectrum attached to an LC-MS peak, you first have to click the row of selected peak followed by selecting the attached spectrum in the plot spectra window.
The method can easily combined with an untargeted Top N approach selecting the top N most intense mz signals of a MS level 1 spectrum for fragmentation.
[5]:
%%capture
# 1. we perform ms level one feature detection
t = emzed.run_feature_finder_metabo(
peakmap_ms2, verbose=False, **ff_params
)
# 2. We attach existing ms2 spectra to corresponding LC-MS level1 peaks detected
emzed.ms2.attach_ms2_spectra(t, mz_tolerance=0.003)
# 3. We filter for all peaks with attached spectra
t = t.filter(t.spectra_ms2.is_not_none())
[6]:
t
[6]:
| id | feature_id | feature_size | mz | mzmin | mzmax | rt | rtmin | rtmax | intensity | quality | fwhm | z | source | spectra_ms2 | num_spectra_ms2 | ms2_extraction_info |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str | object | int | str |
| 31 | 9 | 2 | 201.048024 | 201.047974 | 201.048141 | 0.50 m | 0.48 m | 0.54 m | 1.56e+08 | 1.50e-02 | 0.03 m | 1 | ms_prm.mzML | [ |
16 | union |
| 90 | 33 | 4 | 465.096264 | 465.095917 | 465.098541 | 1.77 m | 1.68 m | 2.00 m | 5.18e+07 | 5.70e-03 | 0.06 m | 1 | ms_prm.mzML | [ |
46 | union |
| 420 | 202 | 2 | 524.136726 | 524.135803 | 524.142456 | 1.49 m | 1.43 m | 1.56 m | 4.04e+06 | 4.49e-04 | 0.05 m | 1 | ms_prm.mzML | [ |
12 | union |
| 731 | 393 | 1 | 201.048056 | 201.046951 | 201.048492 | 0.57 m | 0.54 m | 1.32 m | 1.69e+06 | 1.56e-04 | 0.07 m | 0 | ms_prm.mzML | [ |
114 | union |
| 746 | 403 | 2 | 346.089733 | 346.088837 | 346.092896 | 1.16 m | 1.12 m | 1.23 m | 1.53e+06 | 1.51e-04 | 0.04 m | 1 | ms_prm.mzML | [ |
7 | union |
In principle, three lines of code are sufficient to obtain a table with all LC-MS peaks.
In addition emzed module ms2 features exporting .MS files to perform fragment analysis with SIRIUS. The function export_sirius_files.
export_sirius_files(peak_table, output_folder, abs_min_intensity=0.0, rel_min_intensity=0.0, *, overwrite=False)
with arguments
peak_table: peak table with column spectra_ms2 ( added with functionattach_ms_spectras, see above). Columns rt and id can be helpful to identify results in sirius, but are optional.
output_folder: saving location sirius files given as absolute path.
abs_min_intensity: Filters ms2 spectra by absolute minimal intensity. If rel_min_intensity is also specified, only peaks compliant to both criteria are exported. Default value is 0.0.
rel_min_intensity: Filters ms2 spectra by minimal intensity relative to the highest peak. If abs_min_intensity is also specified, only peaks compliant to both criteria are exported. Default value is 0.0
overwrite: If True existing .ms files in output_folder can be overwritten. Default value is False.
We can apply the function export_sirius_files directly to the table t we just created in above paragraph:
[7]:
# we use the local path of this script as output folder
here = os.getcwd()
# we apply the function without filtering
emzed.ms2.export_sirius_files(t, here, overwrite=True)
# we have a look into output_folder
from glob import glob
paths = glob("*.ms")
[os.path.basename(p) for p in paths]
# we load the first spectra
with open(paths[3], "r") as fp:
x = fp.read()
print(x[:500])
# we determine for the number of lines
len(x.splitlines())
>compound 90
>rt 106.446684
>parentmass 465.09722900390625
>ms2
51.05709457397461 6327.0234375
51.367069244384766 8147.32470703125
53.03940200805664 128142.3671875
55.018497467041016 128520.65625
57.0341796875 207778.125
57.53778839111328 5615.81982421875
57.9855842590332 10198.4140625
57.987945556640625 66420.40625
58.99308395385742 13428.9453125
58.995731353759766 112538.6015625
61.02908706665039 148800.203125
66.23013305664062 6748.08349609375
66.2993392944336 20918.775390625
66.3019180297851
[7]:
111
Hence export_sirius_files creates an .ms file for each row in table t containing ms level 2 spectra. The resulting .ms file has the entries compound, rt, parentmass and ms2, where compound and rt are optional. Since by default no filtering is applied, the resulting .ms file contains a huge number of spectra, where most peaks are noise. In particular merging spectra with method union increases the number of noise peaks and a meaningful analysis of the
fragmentation pattern is massively hampered. Therefore, we repeat the export with filtering by introducing a minimal peak intensity relative to the most intense peak with argument rel_min_intensity:
[8]:
emzed.ms2.export_sirius_files(
t, here, rel_min_intensity=0.01, overwrite=True
)
# we have a look into output_folder
from glob import glob
paths = glob("*.ms")
[os.path.basename(p) for p in paths]
# we load the first spectra
with open(paths[2], "r") as fp:
x = fp.read()
print(x)
# we determine for the number of lines
len(x.splitlines())
>compound 31
>rt 30.0295662
>parentmass 201.04805755615234
>ms2
201.04794311523438 500875360.0
[8]:
5
Filtering massively reduced the number of spectral peaks from 111 to 19.
Attaching MS level 2 spectra works smoothly with ms2 module. In principle, we can perform the same operation using PeakMap methods introduced in chapter 7. Let’s start again with exploring the peak map:
As we have already seen, the example peak map contains 4 different precursor ions. To get the specific ms2 data of each precursor we can use the split_by_precursors() method:
[9]:
prec2pm = peakmap_ms2.split_by_precursors()
prec2pm
[9]:
{(524.138,): <PeakMap 2623468021eedb6ea9de54ff1c04e782>,
(465.098,): <PeakMap 7c7f25c67abedb60d8c0e35054b3d72d>,
(346.09,): <PeakMap c68aca7ec0248b6cdcae05831e500b40>,
(201.048,): <PeakMap 1d6445c8f47d86a9b0033cbde9bfea8a>}
We obtain a dictionary with the precursor as key and the corresponding PeakMap as value. Let’s plot the corresponding TIC of PeakMaP with the first precursor ion 465.098
[10]:
import matplotlib.pyplot as plt
precursor_mz = (465.098,)
pm_465 = prec2pm[precursor_mz]
chromatogram = pm_465.chromatogram()
# we plot the chromatogram
plt.plot(
*chromatogram
) # Note: chromatogram is a tuple with the 2 arrays 'RT values' and 'intensities'
plt.xlabel("Retention time (sec)")
plt.ylabel("Intensity")
plt.title("MS2 TIC for precursor %s m/z" % precursor_mz)
plt.show()

We observe the major peak eluting between 100 and 120 sec with apex at approximately 110 sec. Next we will build the corresponding consensus spectrum. There are several ways to extract the spectra within retention time window. We can use PeakMap methods filter or extract:
[11]:
spectra = pm_465.filter(lambda v: 100 <= v.rt <= 120).spectra
len(spectra)
[11]:
47
[12]:
spectra = pm_465.extract(rtmin=100, rtmax=120).spectra
len(spectra)
[12]:
47
Finally we produce an averaged MS2 spectrum of all selected spectra. To do so, we can use the emzed.ms2.merge_spectra function:
emzed.ms2.merge_spectra(spectra, mz_binning_width, is_ppm=False)
with arguments:
spectra: list of Spectrum objects or PeakMap.spectra attribute of same MS level
mz_binning_width: absolute (Da) or relative (ppm) m/z tolerance parameter for matching m/z values
is_ppm: indicates if mz_binning_width is in Da (absolute) or ppm (relative). Default value is False
[13]:
averaged_spectrum = emzed.ms2.merge_spectra(
spectra, mz_binning_width=5.0, is_ppm=True
)
plt.stem(
averaged_spectrum.mzs,
averaged_spectrum.intensities,
markerfmt=" ",
basefmt="C0-",
)
plt.xlim((100, 480))
plt.xlabel("m/z")
plt.ylabel("Intensity")
plt.show()
len(averaged_spectrum.peaks)

[13]:
127
We can now setup a quick routine resulting a table that contains all prescursors with corresponding averaged spectra:
[14]:
def build_average_spectrum(pm, bin_width, is_ppm):
merge = emzed.ms2.merge_spectra
return [merge(spectra, mz_binning_width=bin_width, is_ppm=is_ppm)]
def get_ms2spectra(peakmap, bin_width=5.0, is_ppm=True):
prec2pm = peakmap.split_by_precursors()
precursors, peakmaps = zip(*prec2pm.items())
t = emzed.to_table("precursor_mz", precursors, object)
t.add_column("peakmap", peakmaps, emzed.PeakMap)
t.add_column(
"spectra_ms2",
t.apply(build_average_spectrum, t.peakmap, bin_width, is_ppm),
object,
)
t.add_enumeration()
return t
[15]:
get_ms2spectra(peakmap_ms2)
[15]:
| id | precursor_mz | spectra_ms2 |
|---|---|---|
| int | object | object |
| 0 | (524.138,) | [ |
| 1 | (465.098,) | [ |
| 2 | (346.09,) | [ |
| 3 | (201.048,) | [ |
Let us use the resulting table to build .ms files for sirius.
At first glance, the second approach seems to be more cumbersome. However, it allows further customizing the procedure and testing new strategies. An example. The method export_sirius_files only allows filtering exported spectra by their relative or absolute intensities. Of course, other filtering methods could be useful and we could perform a filter step prior to exporting the spectra.
In the following, we introduce a concept that allows filtering ms2 spectra attached to a peak table. We can define a general filter function:
[16]:
def filter_spectra(spectra, filter_function, kwargs):
return [
filter_function(spectrum, **kwargs) for spectrum in spectra
]
As an example, we define a filter function returning a Spectrum with n most intense fragment ions:
[17]:
def get_top_n_spectrum(spectrum, top_n=5):
# we extract the top n peaks from the spectrum.peaks array
s = spectrum
peaks = s.peaks
# 1. we sort the peaks by intensities
ascending_peaks = peaks[peaks[:, 1].argsort()]
# 2. we check the number of available spectra
top_n = top_n if len(peaks) >= top_n else len(peaks)
# 3. we extract the n most intense peaks
top_n = ascending_peaks[-top_n:]
top_n.sort() # we sort the peaks again by increasing m/z values
# we create a new spectrum object
return emzed.Spectrum(
s.scan_number,
s.rt,
s.ms_level,
s.polarity,
s.precursors,
top_n,
)
We can now apply the filter function get_top_n_spectrum to our table t using the general filter function filter_spectra:
[18]:
kwargs = {"top_n": 6}
function_arguments = [t.spectra_ms2, get_top_n_spectrum, kwargs]
t.replace_column(
"spectra_ms2",
t.apply(filter_spectra, *function_arguments),
object,
)
# We check whether all spectra contain 6 or less peaks
# each cell of colum 'spectra_ms2' contains a list with a single spectrum
all([len(s[0].peaks) <= 6 for s in t.spectra_ms2])
[18]:
True
Note, by separating list comprehension and spectral filtering, the filter function get_top_n_spectrum can be easier applied in other context, which is in line with the concept “a function should have only one function”. The example also demonstrates the strength of emzed framework for testing new / customized processing steps.

emzed run_feature_finder_metabo supports feature detection of ms level 2 peaks. Given example peak map contains chromatograms of MS level 1 and 2 acquired with an Thermo QExactive instrument. We can apply run_feature_finder_metabo to ms level 2 data.
[19]:
%%capture
arguments = {
"common_chrom_fwhm": 5.0,
"mtd_noise_threshold_int": 100,
"epdet_min_fwhm": 2.0,
"epdet_max_fwhm": 30.0,
"ms_level": 2,
"split_by_precursors_mz_tol": 0.0,
}
t = emzed.run_feature_finder_metabo(peakmap_ms2, **arguments)
t[:5]
To handle feature detection on MS level 2 run_feature_finder_metabo extracts LC-MS peaks precursor-wise by default.
Similar, emzed.quantification.integrate supports MS level 2 peak integration:
[20]:
t1 = emzed.quantification.integrate(
t, "linear", ms_level=2, show_progress=False
)
[21]:
t1[:3]
[21]:
| id | feature_id | feature_size | mz | mzmin | mzmax | rt | rtmin | rtmax | intensity | quality | fwhm | z | source | precursors | peak_shape_model | area | rmse | valid_model |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int | int | int | MzType | MzType | MzType | RtType | RtType | RtType | float | float | RtType | int | str | object | str | float | float | bool |
| 0 | 0 | 1 | 201.047957 | 201.047546 | 201.051926 | 0.50 m | 0.46 m | 1.50 m | 9.85e+07 | 9.99e-01 | 0.03 m | 0 | ms_prm.mzML | (201.048,) | linear | 1.11e+08 | 0.00e+00 | True |
| 1 | 1 | 1 | 217.042881 | 217.042603 | 217.043259 | 1.78 m | 1.68 m | 2.00 m | 3.27e+07 | 5.90e-01 | 0.06 m | 0 | ms_prm.mzML | (465.098,) | linear | 4.33e+07 | 0.00e+00 | True |
| 2 | 2 | 1 | 160.075640 | 160.075500 | 160.075928 | 1.49 m | 1.44 m | 1.54 m | 9.32e+05 | 4.98e-01 | 0.07 m | 0 | ms_prm.mzML | (524.138,) | linear | 9.57e+05 | 0.00e+00 | True |
© Copyright 2012-2025 ETH Zurich
Last build 2025-10-28 13:23:38.200852.
Created using Sphinx 8.1.3.